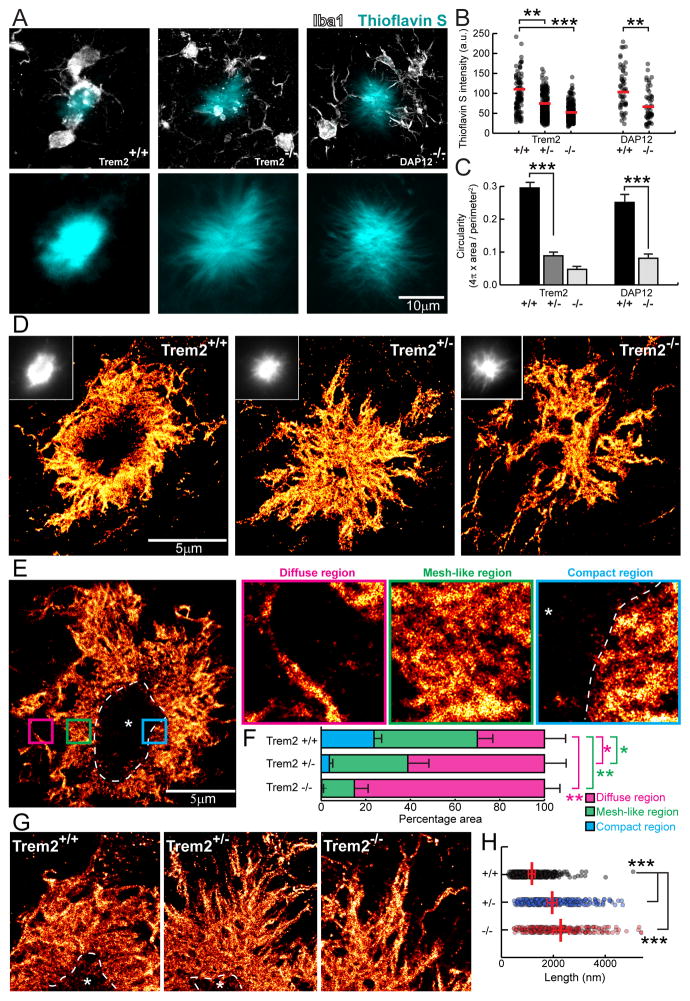
Figure 3. Super-resolution STORM imaging reveals a marked reduction in plaque compaction in Trem2 haplodeficiency.
(A) Confocal images of thioflavin S-labeled amyloid plaques in Trem2+/+ and Trem2−/− 5XFAD mice and DAP12−/− APPPS1-21 mice. (B and C) Quantification of mean thioflavin S fluorescence intensity and circularity (roundness) in plaques from 5XFAD mice with different Trem2 genotypes and APPPS1-21 mice with different DAP12 genotypes. N=3 mice for each group, 747 plaques for 5XFAD:Trem2 and 198 plaques for APPPS1-21:DAP12. (D and E) Immunolabeled Aβ plaque fibrils reconstructed by super-resolution STORM imaging. Images were pseudo-colored according to the reconstructed intensities. Insets in D show the same plaque imaged with wild-field illumination. Dashed line and asterisk in E indicate locations of the compact plaque core which is not labeled due to poor antibody penetration in 5XFAD Trem2+/+ mice (Figure S6). Three insets in E show high zoom examples of different types of Aβ fibril organization located in plaque regions with different degree of compaction. (F) Quantification of the proportion of diffuse, mesh-like and compact regions in amyloid plaques from 5XFAD mice with different Trem2 copy numbers. N=8 plaques from each genotype. Multiple t-tests were used to compare different regions between genotypes. Multiple comparisons were corrected with Holm-Sidak method. *: p<0.0005, **: p<0.0001. (G) Example images of fibrils within the diffuse plaque region extending radially from the plaque center in 5XFAD mice with different Trem2 genotypes. Dashed lines and asterisks indicate locations of the unlabeled compact plaque core. (H) Quantification of lengths of the diffuse fibrils in plaques of 5XFAD mice with different Trem2 copy numbers. Scatter plot shows individual data point. Red bars indicate group averages by genotypes. Except for comparison in F, Student t-tests were used for all statistical comparisons, **: p<0.01, ***: p<0.001; a.u.: arbitrary unit.