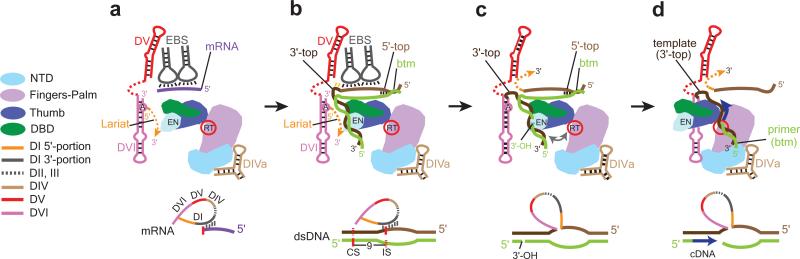
Fig. 8. Structure-based Model for Steps from RNA Splicing to TPRT.
(a) Post-RNA splicing state observed in our RNP structure. The RNP in the post-catalytic state is shown with mRNA bound to the EBS's, stabilized by the LtrA thumb domain. The solid magenta line represents the mRNA captured in the RNP reconstruction. (b) DNA (shown in shades of brown and green) binding to the RNP complex. The top strand of DNA IBS's (lighter brown) base-pair with intron EBS's after displacement of the mRNA. A bend is shown in the DNA. Orientations of the intron RNP and DNA are speculative. (c) Reverse splicing and bottom-strand cleavage. After RNA integration into the insertion site (IS) on DNA, the bottom strand (green) is cleaved 9 nt 3’ to the IS generating the 3’-OH that primes the TPRT reaction. The distance between the endonuclease and RT active sites is depicted by a bidirectional arrow. (d) Trimolecular RNP complex poised for TPRT. Remodeling of the RNP-DNA complex is required to bring the 3’-OH of the cleaved DNA from the EN to the RT active site.