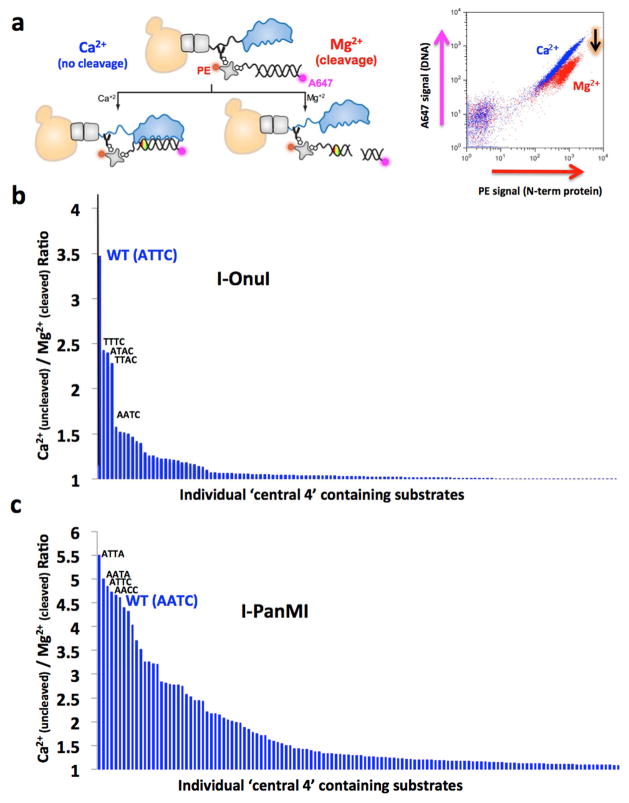
Figure 5.
Analysis of central four specificity for I-OnuI and I-PanMI using the tethered DNA flow cytometric cleavage assay. (A) Schematic of the tethered DNA cleavage assay. Details of the method are illustrated in Figures S4 and S5 and further described in Materials and Methods and in the Supplemental Experimental Procedures. (B) Ranked array of cleavage activity by I-OnuI against DNA target substrates containing basepair combinations spanning positions −2 to +2. Only three substrates are cleavable at an efficiency of ≥50% that of the wild-type central four target; the remainder are very inefficiently cleaved in this assay. (C) The same type of ranked array of cleavage activity of DNA substrates by the I-PanMI enzyme. Note the much broader tolerance of variation in the central four target sequence. See also Figure S6 for the raw flow cleavage assay data for the I-PanMI enzyme.