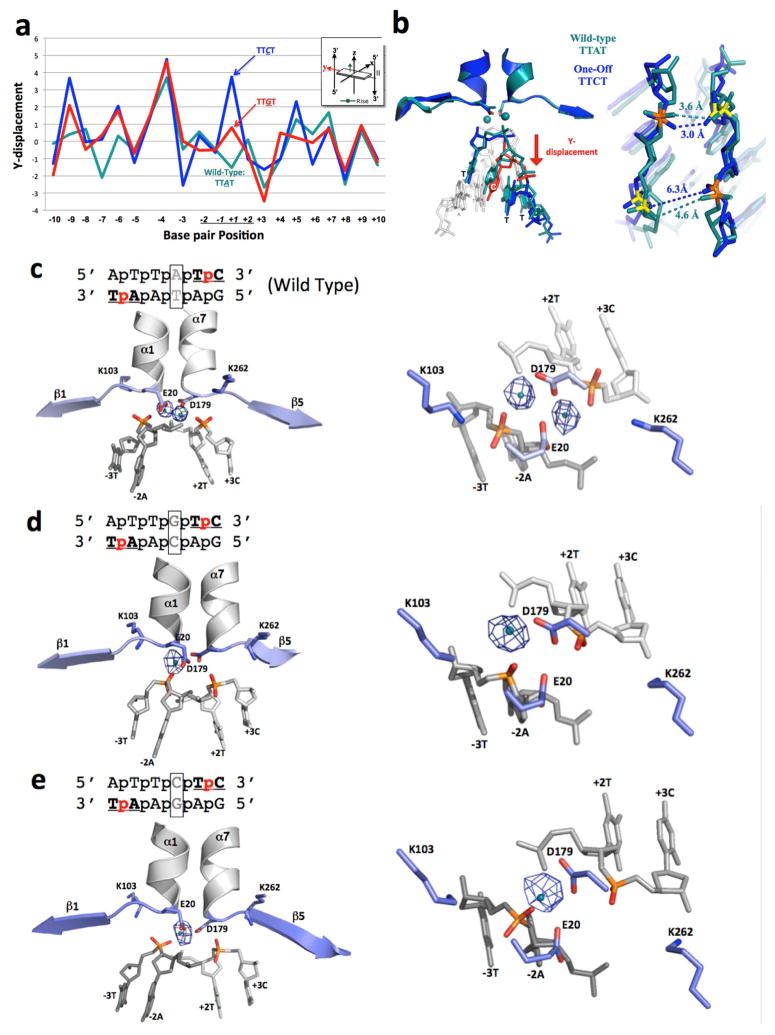
Figure 6.
Structural analysis of I-SmaMI bound to its wild-type DNA target (which contains a ‘TTAT’ central four sequence) and to two variant DNA targets that each contain a single basepair substitution in the central four region (corresponding to ‘TTCT’ and ‘TTGT’). (A) 3DNA analysis of ‘Y-displacement’ across the three DNA target sites. Note the movement of the substituted basepair in the two variant target sites. See also Figure S7 for the same analysis for ‘twist’ and ‘roll’ parameters. (B) Superposition of the central four DNA region of the wild-type (‘TTAT’) structure (teal) and the variant ‘TTCT’ structure (blue). Distances between calcium-coordinating phosphate oxygens are indicated in angstroms. (C)–(E): Anomalous difference Fourier maps for I-SmaMI bound to wild-type and variant target sites, shown from the side (left) and looking down at the active site from above (right). Anomalous electron density from the bound calcium is shown as blue mesh (using a contour level cutoff of 5.0). See also Figure S8 for data demonstrating the ability of Mn2+ to rescue cleavage of the ‘TTAT’ and ‘TTCT’ containing substrates by the same enzyme.