Abstract
Neuropathic pain often develops following nerve injury as a result of maladaptive changes that occur in the injured nerve and along the nociceptive pathways of the peripheral and central nervous systems. Multiple cellular and molecular mechanisms likely account for these changes; however, the exact nature of these mechanisms remain largely unknown. A growing number of studies suggest that alteration in gene expression is an important step in the progression from acute to chronic pain states and epigenetic regulation has been proposed to drive this change in gene expression. In this review, we discuss recent evidence that the DNA-binding protein Neuron-Restrictive Silencing Factor/Repressor Element-1 Silencing Transcription factor NRSF/REST) is an important component in the development and maintenance of neuropathic pain through its role as a transcriptional regulator for a select subset of genes that it normally represses during development.
Keywords: Epigenetics, Neuropathic pain, NRSF/REST, MeCP2
Introduction
Neuropathic pain is a persistent nociceptive hypersensitivity caused by either injury or disease to the somatosensory nervous system. It is widely considered to be one of the most difficult pain syndromes to treat, and it creates a substantial economic burden for modern healthcare [37]. While a precise number of affected individuals is difficult to determine given the variable and subjective nature of the symptoms, a recent systematic review of epidemiological studies of neuropathic pain in the general population estimates the population prevalence of pain with neuropathic characteristics between 6.9% and 10% [61]. The mechanisms responsible for the transition from acute to chronic pain following neural injury are not well understood, but this transition is accompanied by significant changes in the transcriptome of both the central and peripheral circuits [1, 19]. These long-term changes in gene expression profiles are mediated, in part, by epigenetic mechanisms that reorganize chromatin structure. The Neuron-Restrictive Silencing Factor/Repressor Element-1 Silencing Transcription factor NRSF/REST) is a DNA-binding protein that represses neuronal gene transcription by recruiting several chromatin-modifying co-factors. Several studies have shown that NRSF-mediated repression of specific genes is an important component in the development and maintenance of neuropathic pain. This review discusses this role of NRSF in neuropathic pain as a transcriptional regulator for a select subset of genes that it normally represses during development.
Neuropathic Pain
The primary hallmark of chronic pain is persistent nociceptive hypersensitivity resulting in a significant reduction in the thresholds required to induce a painful response [23, 63, 69]. This typically manifests as either allodynia, where a previously innocuous stimuli now causes pain, or hyperalgesia, which results in an amplified response to a noxious stimuli [64]. Chronic pain often develops following acute pain as the result of tissue damage and persistent inflammation or pathological adaptive plasticity in the peripheral and central nervous systems. This chronic pain is classified as neuropathic when it arises as a direct result of damage or disease to the somatosensory system. There is considerable variability in which individuals will transition from an acute pain state to a chronic pain state, and the mechanisms underlying this variability are largely unknown. Given this variability and a lack of understanding about its causes, it should come as no surprise that current therapeutic strategies aimed at treating chronic pain are largely ineffective, with over half of chronic pain patients having little or no control over their pain using currently available therapies [31].
The development and subsequent maintenance of chronic pain states involves persistent changes to multiple regions within both the peripheral and central nervous system. These alterations along the nociceptive pathway occur at both the cellular and molecular levels. For example, neurons in the dorsal horn exhibit increased excitatory responses as well as decreased firing thresholds [20]. These aberrant neuronal activities and the development of neuropathic pain behaviors correlate with changes in gene expression along the entire length of the pain transduction and modulation pathways, including supraspinal, spinal, and brain regions (reviewed in [15]). The mechanism(s) that drive these changes in gene expression, which persists for long periods well after the precipitating injury has resolved, are poorly understood. Recently, interest has turned to epigenetic mechanisms as a possible means of driving these transcriptome changes. Epigenetic changes are well known to modify neuronal activity in synaptic plasticity, learning, and memory. In addition, pathophysiologic changes in a number of neuropsychiatiric disorders involve epigenetic modifications [5]. Recent studies have demonstrated epigenetic changes in both spinal and brain regions in chronic pain disorders, contributing to sensitization to thermal and mechanical stimuli [21, 68]. Taken together, these studies suggest that epigenetic mechanisms are an important mediator of the development and maintenance of neuropathic pain. To date, however, few studies have identified specific players critical to driving these epigenetic changes.
REST/NRSF
NRSF is a zinc finger DNA-binding protein that mediates both transient and long-term repression of neuronal gene transcription. A bipartite interaction domain within NRSF recruits the SIN3A/B and CoREST co-repressor complexes through its N- and C-terminal regions, respectively. These co-repressor complexes epigenetically modify target genomic regions through several chromatin-modifying enzymes, including Histone Deacetylases 1/2 (HDAC1/2), the histone demethylase LSD1, and the histone methylase G9a (reviewed in [39]). The CoREST complex also contains the ATP-dependent helicase BRG1, which improves NRSF access to the target gene regions [38]. Recruitment of CoREST can also be stabilized by interactions with methyl-CpG-binding protein (MeCP2) [4]. Recent studies suggest that NRSF-mediated silencing of gene transcription is also assisted by the recruitment of Polycomb Repressive Complexes through interactions between the subunits of the Polycomb and NRSF protein complexes [16, 33, 46, 58].
Initially considered a developmental master regulator of neuronal cell fate [13, 51], NRSF is now recognized as having an important regulatory role in several neural diseases as well (recently reviewed in [56]). During development, NRSF is expressed at high levels in non-neuronal tissues and suppresses neuronal specification by repressing neuronal gene transcription [13, 51]. Although the loss of NRSF can derepress transcription of some target genes, the lack of NRSF does not alter cell fate specification of non-neuronal tissue [11]. Compared to non-neuronal tissue, NRSF is expressed at lower levels in both embryonic and adult neural progenitor cells and its expression level diminishes as differentiation proceeds [4, 17]. NRSF remains expressed at low levels, however, within mature neurons of several brain regions [32, 43]. NRSF has multiple alternatively spliced isoforms including a neuron specific isoform, REST4, which weakly binds DNA [27, 43]. The full functional significance of the truncated REST4 isoform in neurons, however, is not completely understood.
In contrast to the maintenance of low NRSF expression levels, NRSF expression is upregulated under some pathological conditions in the adult nervous system. Global ischemic stroke in rodents, for example, elevates mRNA and protein levels of both NRSF and CoREST in hippocampal CA1 neurons [8, 36]. This up-regulation represses a battery of genes essential for synaptic function and plays a causal role in the death of CA1 neurons following stroke. High NRSF expression levels also characterize several neural-lineage cancers, such as medulloblastomas and glioblastomas. Disruption of NRSF-mediated repression in these cancers leads to apoptosis and a reduction in tumor size [24, 26], suggesting that NRSF promotes self-renewal of neural tumor cells in a manner similar to its role in maintaining neural progenitor pools during development.
Pain-induced transcriptional repression mediated by NRSF
In addition to stroke and cancer, NRSF expression levels are upregulated in neuropathic pain. Remifentanil is a short-acting synthetic opioid that is widely used as a general anesthetic, but can also induce post-operative hyperalegesia. Infusion of remifentanil in an incisional post-operative pain mouse model elevated NRSF expression levels in the periaqueductal gray [29]. This increase in NRSF was associated with a decrease in paw withdraw times in response to mechanical and thermal stimulation. Consistent with studies in cultured cells showing that transcription of Oprm1 (encoding the μ-opioid receptor) is directly repressed by NRSF [25], Oprm1 expression in the periaqueductal gray was repressed by remifentanil-induced elevation of NRSF and ventricular administration of antisense oligonucleotide directed against NRSF rescued both Oprm1 expression levels and paw withdrawal latency times.
Pain is also associated with an upregulation of NRSF expression in the peripheral nervous system. Studies using partial sciatic nerve ligation in mice showed an increase in both NRSF mRNA and protein levels in dorsal root ganglia that was maintained for several weeks following injury [47, 59, 60]. Expression levels for the NRSF target genes Kcnd3, Kcnq2 and Scn10a (which respectively encode the Kv 4.3, Kv 7.2 and Nav1.8 channels) as well as Oprm1 were repressed in response to the increase in NRSF. Repression of these target genes was associated with both persistent C-fiber dysfunction and disruption of K+ M-type currents that facilitated neuropathic excitability of peripheral sensory fibers. Knockdown of NRSF with antisense oligonucleotides was sufficient to rescue both expression levels for several of these NRSF target genes and C-fiber function [59].
Consistent with a role for chromatin remodeling in the development of persistent pain, administration of histone deacetylase (HDAC) inhibitors can alleviate pain symptoms in several model systems (reviewed in [3]). For NRSF target genes, such Kcnd3 and Scn10a, persistent pain is associated with hypoacetylation of genomic regions surrounding the NRSF binding sites [59, 60] which suggests that HDAC inhibitors ameliorate neuropathic pain, at least in part, by counteracting chromatin remodeling and transcriptional repression mediated by NRSF. Given the ability to HDAC inhibitors to alleviate pain by restoring expression levels of NRSF target genes, we have examined whether Glutamic acid decarboxylase 2 (Gad2) expression in the raphe magnus is regulated by NRSF. GABAergic interneurons in the raphe magnus are integral to suppressing descending facilitation of pain [12]. In rodents, both inflammatory pain and spinal nerve ligation-induced neuropathic pain models decrease histone acetylation levels at the Gad2 promoter and repress Gad2 expression levels [68]. This repression is also associated with the recruitment of HDAC 1/2 enzymes to the Gad2 promoter. Since these HDAC enzymes are part of the co-repressor protein complexes recruited by NRSF, the Gad2 promoter nucleotide sequence was examined for NRSF binding sites. Our analysis revealed an NRSF consensus binding site that was highly conserved in mammals, and the ENCODE database confirms that this Gad2 promoter region is bound by NRSF and its co-repressors SIN3A and HDAC2. Together, these findings suggest that NRSF directly binding to the Gad2 promoter may be involved in the repression of Gad2 seen in neuropathic pain states.
Selective targets gene repression by NRSF in neuropathic pain
The full set of genes directly targeted by NRSF that are involved in neuropathic pain has not yet been established. Bioinformatic analyses using consensus NRSF target sequences have identified putative NRSF binding sites within the genomes of several vertebrate species [6, 52], and high throughput sequencing of immunoprecipitated genomic DNA has shown which putative binding sites are occupied in different cell types [40, 50]. The NRSF ChIP-seq datasets include a series of studies with several human cell lines that are incorporated into the ENCODE database [14].
Comparing genomic regions occupied by NRSF in the ENCODE database to a meta-analyses of microarray and RNA-seq studies in neuropathic pain mouse models [44] reveals several additional genes that are likely direct targets of NRSF-mediated repression. These down-regulated genes include several neurotransmitter receptors subunits (Chrna3, Gabbr1, Grik1 and Htr3a), the Kcnc2 voltage-gated potassium channel and the Snap25 synaptosomal-associated protein. The ENCODE database shows that NRSF associates with these genes either near their transcription start site or within introns, and that these regions are also typically occupied by many NRSF cofactors including SIN3A, CoREST and HDAC1/2.
Elevated levels of NRSF, however, are not likely sufficient to account for the repression of target genes in neuropathic pain conditions. Many genes upregulated in the microarray and RNA-seq studies of neuropathic pain mouse models also have promoter regions that show occupancy by NRSF in the ENCODE database. These upregulated genes include Gabra5 (GABAa receptor α5 subunit), Cacna2d1 (an L-type voltage-gated calcium channel), Vip (Vasoactive intestinal peptide), Gadd45a (DNA demethylase) and Gap43 (growth associated protein 43). The ENCODE database also shows that these up-regulated genes are occupied many NRSF cofactors, including SIN3A, CoREST and HDAC1/2. Since most cell types in the ENCODE are non-neural, the genes associated with NRSF in the database are more likely to be representative of the genes targeted by NRSF during development. Thus, the comparison of microarray and RNA-seq studies in neuropathic pain mouse models indicates that NRSF selectively targets only a subset of the genes it represses during development.
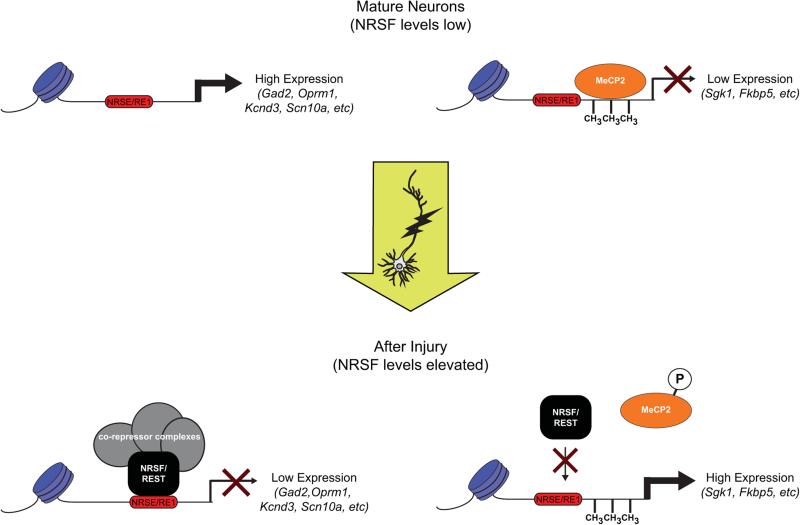
Selectivity in target gene repression suggests other factors modulate NRSF's ability to either bind target gene regions or recruit co-repressors. Pain-induced phosphorylation of MeCP2 is one potential mechanism to modify repression by NRSF. MeCP2 is a methylated DNA-binding protein that recruits co-repressor complexes containing HDACs (reviewed in [2]), and, as noted above, MeCP2 can facilitate recruitment of CoREST to NRSF [30]. Phosphorylated MeCP2, however, has a diminished affinity for methylated DNA, and MeCP2 target gene transcription levels are derepressed when MeCP2 is phosphorylated [10]. In an inflammatory pain mouse model, phosphorylated MeCP2 levels were elevated in subsets of dorsal horn neurons, and the expression of MeCP2 target genes, including Sgk1 and Fkbp5, was increased [18]. Examination of the ENCODE database reveals that respective promoter regions for Sgk1 and Fkbp5 are also targeted by NRSF and CoREST. This suggests the possibility that phospho-MeCP2 disrupts NRSF repression of genes that these factors co-repress. Thus, future studies are needed to establish why the NRSE on some genes is insufficient to mediate gene repression when MeCP2 is phosphorylated despite the fact that NRSF is able to repress other target genes under these conditions (Figure 1).
Figure 1. Differential Responses of NRSF Target Genes.
In mature neurons, many NRSF target gene expression levels are elevated because of NRSF is present at only low levels. Some NRSF target genes, however, are repressed in mature neurons due to the action of other repressor proteins, such MeCP2 binding to methylated DNA. Upon nerve injury, NRSF levels are elevated and transcription of several NRSF target genes (e.g., Gad2, Oprm1, Kcnd3, Scn10a) is repressed due to increased NRSF occupancy at NRSF binding sites (NRSE/RE1). For some NRSF-responsive genes that are also regulated by MeCP2 (e.g., Sgk1, Fkbp5), however, nerve injury derepresses expression levels. MeCP2 is phosphorylated following nerve injury, which reduces its affinity for methylated DNA. Future studies are needed to establish why the NRSE on some genes is insufficient to mediate gene repression when MeCP2 is phosphorylated after injury, despite the fact that NRSF is able to repress other target genes under these conditions.
In addition to protein coding genes, expression levels for several micro-RNAs (miRNAs) are downregulated under chronic pain conditions (reviewed in [15]). Expression levels for miR-7a, 96 and 134 were reduced in the dorsal root ganglia of various rodent pain model systems [9, 34, 48]. None of the promoter regions for these downregulated miRNAs, however, have either consensus NRSF binding site sequences or association with NRSF binding in the ENCODE database. This suggests that the repression of these miRNAs during neuropathic pain is not mediated by NRSF, although it is possible that distant NRSF complexes could modulate expression of these miRNAs. In development, by contrast, NRSF represses miR-21 expression in embryonic stem cells [55]. Interestingly, miR-21 expression is elevated in the dorsal root ganglia for at least two weeks in rats after either spinal nerve ligation or chronic constriction injury [49]. This pain-induced increase in miR-21 expression is further evidence that, under neuropathic pain conditions, NRSF represses only a select subset of its developmental target genes.
Future Considerations
Neuropathic pain is associated with an increase in NRSF levels in both central and peripheral pain circuits, and this elevation of NRSF selectively represses a set of target genes within the neurons of these circuits. A better understanding of NRSF's role in neuropathic pain will require establishing the mechanisms that initiate and maintain the upregulation of NRSF expression following neural injury as well as the mechanisms that guide NRSF to repress a select subset of target genes.
There are several non-mutually exclusive potential mechanisms to upregulate NRSF expression levels. The Wnt signaling pathway activates Nrsf transcription in chick embryos [35], and several studies have linked neuropathic pain to Wnt pathway activation [22, 53, 54, 66, 67]. Thus, establishing whether the Wnt pathway directly activates Nrsf expression will significantly improve our understanding the dynamics of injury-induced Nrsf transcription. In addition to transcriptional regulation, the contribution of post-transcriptional regulatory mechanisms, including alternative splicing and miRNA regulation, remain to be clarified. As noted above, NRSF has several alternatively spliced isoforms, including a truncated REST4 isoform that weakly binds to DNA. The balance between full length NRSF and REST4 isoforms appears to be important for the maintenance and differentiation of neural progenitor cells [41, 45]. Studies in epilepsy and fetal alcohol exposure rodent models have respectively reported that the full-length NRSF and REST4 isoform ratio contributes to the homeostatsis of neural networks and neuroprotection [7, 57]. Whether this isoform ratio is altered in neuropathic pain and if such a change is integral to the maladaptive in either peripheral or central pain circuits remains to be established. Finally, our knowledge of how NRSF levels are regulated by miRNA following neural injury is limited. Several miRNAs have been reported to target NRSF, both directly and indirectly, and for several of these miRNAs, NRSF mediates reciprocal repression [28, 42, 62, 65].
Selective repression of subset target genes by NRSF in response to neural injury suggests that other regulatory factors modulate NRSF's ability to either bind target gene regions or recruit co-repressors. As discussed above, interactions with phosphorylated MeCP2 following neural injury are a potential mechanism for a subset of target genes to escape NRSF-mediated repression, but this mechanism remains to be verified. Alternatively, interactions with Polycomb Group complexes [16, 33, 46, 58] may also influence NRSF target gene repression, but the contribution of Polycomb-mediated neuropathic pain is currently poorly understood. Elucidating the intricate regulatory web modulating both NRSF target gene selection and NRSF expression levels are essential to understanding how the epigenetic program controlled by NRSF become maladaptive and facilitate neuropathic pain.
Acknowledgments
NIH NR010797 (DEW), NIH DC008955 (JWC) and the Burke Medical Research Institute Foundation (DEW and JWC).
Abbreviations
- ENCODE
Encyclopedia of DNA elements
- HDAC
histone deacetylase
- MeCP2
methyl CpG binding protein 2
- miRNA
micro-RNA
- NRSF
Neuron-Restrictive Silencing Factor
- REST
Repressor Element-1 Silencing Transcription
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alvarado S, Tajerian M, Millecamps M, Suderman M, Stone LS, Szyf M. Peripheral nerve injury is accompanied by chronic transcriptome-wide changes in the mouse prefrontal cortex. Mol Pain. 2013;9:21. doi: 10.1186/1744-8069-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausio J, de Paz AM, Esteller M. MeCP2: the long trip from a chromatin protein to neurological disorders. Trends Mol Med. 2014;20:487–498. doi: 10.1016/j.molmed.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Bai G, Ren K, Dubner R. Epigenetic regulation of persistent pain. Transl Res. 2015;165:177–199. doi: 10.1016/j.trsl.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Brami-Cherrier K, Anzalone A, Ramos M, Forne I, Macciardi F, Imhof A, Borrelli E. Epigenetic reprogramming of cortical neurons through alteration of dopaminergic circuits. Molecular psychiatry. 2014;19:1193–1200. doi: 10.1038/mp.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai L, Bian M, Liu M, Sheng Z, Suo H, Wang Z, Huang F, Fei J. Ethanol-induced neurodegeneration in NRSF/REST neuronal conditional knockout mice. Neuroscience. 2011;181:196–205. doi: 10.1016/j.neuroscience.2011.02.059. [DOI] [PubMed] [Google Scholar]
- 8.Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, Grooms SY, Regis R, Bennett MV, Zukin RS. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen HP, Zhou W, Kang LM, Yan H, Zhang L, Xu BH, Cai WH. Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury. Neurochem Res. 2014;39:76–83. doi: 10.1007/s11064-013-1192-z. [DOI] [PubMed] [Google Scholar]
- 10.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 11.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 12.Cho HJ, Basbaum AI. GABAergic circuitry in the rostral ventral medulla of the rat and its relationship to descending antinociceptive controls. J Comp Neurol. 1991;303:316–328. doi: 10.1002/cne.903030212. [DOI] [PubMed] [Google Scholar]
- 13.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 14.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Descalzi G, Ikegami D, Ushijima T, Nestler EJ, Zachariou V, Narita M. Epigenetic mechanisms of chronic pain. Trends Neurosci. 2015;38:237–246. doi: 10.1016/j.tins.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N, Rappsilber J, Sodersten E, Hansen K. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 2012;8:e1002494. doi: 10.1371/journal.pgen.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, Hammer RE, Hsieh J. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci. 2011;31:9772–9786. doi: 10.1523/JNEUROSCI.1604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geranton SM, Morenilla-Palao C, Hunt SP. A role for transcriptional repressor methyl-CpG-binding protein 2 and plasticity-related gene serum- and glucocorticoid inducible kinase 1 in the induction of inflammatory pain states. J Neurosci. 2007;27:6163–6173. doi: 10.1523/JNEUROSCI.1306-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer P, Banck MS, Amberg R, Wang C, Petznick G, Luo S, Khrebtukova I, Schroth GP, Beyerlein P, Beutler AS. mRNA-seq with agnostic splice site discovery for nervous system transcriptomics tested in chronic pain. Genome Res. 2010;20:847–860. doi: 10.1101/gr.101204.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda H, Stark J, Fischer H, Wagner M, Drdla R, Jager T, Sandkuhler J. Synaptic amplifier of inflammatory pain in the spinal dorsal horn. Science. 2006;312:1659–1662. doi: 10.1126/science.1127233. [DOI] [PubMed] [Google Scholar]
- 21.Imai S, Ikegami D, Yamashita A, Shimizu T, Narita M, Niikura K, Furuya M, Kobayashi Y, Miyashita K, Okutsu D, Kato A, Nakamura A, Araki A, Omi K, Nakamura M, James Okano H, Okano H, Ando T, Takeshima H, Ushijima T, Kuzumaki N, Suzuki T, Narita M. Epigenetic transcriptional activation of monocyte chemotactic protein 3 contributes to long-lasting neuropathic pain. Brain. 2013;136:828–843. doi: 10.1093/brain/aws330. [DOI] [PubMed] [Google Scholar]
- 22.Itokazu T, Hayano Y, Takahashi R, Yamashita T. Involvement of Wnt/beta-catenin signaling in the development of neuropathic pain. Neurosci Res. 2014;79:34–40. doi: 10.1016/j.neures.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Ji RR, Kohno T, Moore KA, Woolf CJ. Central sensitization and LTP: do pain and memory share similar mechanisms? Trends Neurosci. 2003;26:696–705. doi: 10.1016/j.tins.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Kamal MM, Sathyan P, Singh SK, Zinn PO, Marisetty AL, Liang S, Gumin J, El-Mesallamy HO, Suki D, Colman H, Fuller GN, Lang FF, Majumder S. REST regulates oncogenic properties of glioblastoma stem cells. Stem Cells. 2012;30:405–414. doi: 10.1002/stem.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, Loh HH. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J Biol Chem. 2004;279:46464–46473. doi: 10.1074/jbc.M403633200. [DOI] [PubMed] [Google Scholar]
- 26.Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, Rastelli L, Marin Dias Carneiro A, Levin V, Fuller GN, Echelard Y, Majumder S. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6:826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 27.Lee JH, Shimojo M, Chai YG, Hersh LB. Studies on the interaction of REST4 with the cholinergic repressor element-1/neuron restrictive silencer element. Brain Res Mol Brain Res. 2000;80:88–98. doi: 10.1016/s0169-328x(00)00129-7. [DOI] [PubMed] [Google Scholar]
- 28.Liang H, Studach L, Hullinger RL, Xie J, Andrisani OM. Down-regulation of RE-1 silencing transcription factor (REST) in advanced prostate cancer by hypoxia-induced miR-106b~25. Exp Cell Res. 2014;320:188–199. doi: 10.1016/j.yexcr.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, Shi L, Zhang J, Kong M, Liu Y, Zhou Y, Xu L, He J, Ma Z, Gu X. Neuron-restrictive silencer factor in periaqueductal gray contributes to remifentanil-induced postoperative hyperalgesia via repression of the mu-opioid receptor. J Neurol Sci. 2015;352:48–52. doi: 10.1016/j.jns.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 31.I.o. Medicine . Relieving Pain in America: A Blueprint for Transforming Prevention. Care, Education, and Research; Washington (DC): 2011. [Google Scholar]
- 32.Mori N, Mizuno T, Murai K, Nakano I, Yamashita H. Effect of age on the gene expression of neural-restrictive silencing factor NRSF/REST. Neurobiol Aging. 2002;23:255–262. doi: 10.1016/s0197-4580(01)00286-x. [DOI] [PubMed] [Google Scholar]
- 33.Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, Margueron R, Ait-Si-Ali S. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell. 2014;53:277–289. doi: 10.1016/j.molcel.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Ni J, Gao Y, Gong S, Guo S, Hisamitsu T, Jiang X. Regulation of mu-opioid type 1 receptors by microRNA134 in dorsal root ganglion neurons following peripheral inflammation. Eur J Pain. 2013;17:313–323. doi: 10.1002/j.1532-2149.2012.00197.x. [DOI] [PubMed] [Google Scholar]
- 35.Nishihara S, Tsuda L, Ogura T. The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochem Biophys Res Commun. 2003;311:55–63. doi: 10.1016/j.bbrc.2003.09.158. [DOI] [PubMed] [Google Scholar]
- 36.Noh M, Mok Y, Lee S, Kim H, Lee SH, Jin GW, Seo JH, Koo H, Park TH, Lee Y. Novel lower critical solution temperature phase transition materials effectively control osmosis by mild temperature changes. Chem Commun (Camb) 2012;48:3845–3847. doi: 10.1039/c2cc30890h. [DOI] [PubMed] [Google Scholar]
- 37.O'Connor AB. Neuropathic pain: quality-of-life impact, costs and cost effectiveness of therapy. Pharmacoeconomics. 2009;27:95–112. doi: 10.2165/00019053-200927020-00002. [DOI] [PubMed] [Google Scholar]
- 38.Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem. 2006;281:38974–38980. doi: 10.1074/jbc.M605370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 40.Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, Yochum GS, Dunn JJ, Goodman RH, Mandel G. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ovando-Roche P, Yu JS, Testori S, Ho C, Cui W. TRF2-mediated stabilization of hREST4 is critical for the differentiation and maintenance of neural progenitors. Stem Cells. 2014;32:2111–2122. doi: 10.1002/stem.1725. [DOI] [PubMed] [Google Scholar]
- 42.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palm K, Belluardo N, Metsis M, Timmusk T. Neuronal expression of zinc finger transcription factor REST/NRSF/XBR gene. J Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins JR, Antunes-Martins A, Calvo M, Grist J, Rust W, Schmid R, Hildebrandt T, Kohl M, Orengo C, McMahon SB, Bennett DL. A comparison of RNA-seq and exon arrays for whole genome transcription profiling of the L5 spinal nerve transection model of neuropathic pain in the rat. Mol Pain. 2014;10:7. doi: 10.1186/1744-8069-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raj B, O'Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol Cell. 2011;43:843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 46.Ren X, Kerppola TK. REST interacts with Cbx proteins and regulates polycomb repressive complex 1 occupancy at RE1 elements. Mol Cell Biol. 2011;31:2100–2110. doi: 10.1128/MCB.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rose K, Ooi L, Dalle C, Robertson B, Wood IC, Gamper N. Transcriptional repression of the M channel subunit Kv7.2 in chronic nerve injury. Pain. 2011;152:742–754. doi: 10.1016/j.pain.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain. 2013;136:2738–2750. doi: 10.1093/brain/awt191. [DOI] [PubMed] [Google Scholar]
- 49.Sakai A, Suzuki H. Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochem Biophys Res Commun. 2013;435:176–181. doi: 10.1016/j.bbrc.2013.04.089. [DOI] [PubMed] [Google Scholar]
- 50.Satoh J, Kawana N, Yamamoto Y. ChIP-Seq Data Mining: Remarkable Differences in NRSF/REST Target Genes between Human ESC and ESC-Derived Neurons. Bioinform Biol Insights. 2013;7:357–368. doi: 10.4137/BBI.S13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 52.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci U S A. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y, Yuan S, Li B, Wang J, Carlton SM, Chung K, Chung JM, Tang SJ. Regulation of Wnt signaling by nociceptive input in animal models. Mol Pain. 2012;8:47. doi: 10.1186/1744-8069-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simonetti M, Agarwal N, Stosser S, Bali KK, Karaulanov E, Kamble R, Pospisilova B, Kurejova M, Birchmeier W, Niehrs C, Heppenstall P, Kuner R. Wnt-Fzd signaling sensitizes peripheral sensory neurons via distinct noncanonical pathways. Neuron. 2014;83:104–121. doi: 10.1016/j.neuron.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 55.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song Z, Zhao D, Zhao H, Yang L. NRSF: an angel or a devil in neurogenesis and neurological diseases. J Mol Neurosci. 2015;56:131–144. doi: 10.1007/s12031-014-0474-5. [DOI] [PubMed] [Google Scholar]
- 57.Spencer EM, Chandler KE, Haddley K, Howard MR, Hughes D, Belyaev ND, Coulson JM, Stewart JP, Buckley NJ, Kipar A, Walker MC, Quinn JP. Regulation and role of REST and REST4 variants in modulation of gene expression in in vivo and in vitro in epilepsy models. Neurobiol Dis. 2006;24:41–52. doi: 10.1016/j.nbd.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 58.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uchida H, Ma L, Ueda H. Epigenetic gene silencing underlies C-fiber dysfunctions in neuropathic pain. J Neurosci. 2010;30:4806–4814. doi: 10.1523/JNEUROSCI.5541-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchida H, Sasaki K, Ma L, Ueda H. Neuron-restrictive silencer factor causes epigenetic silencing of Kv4.3 gene after peripheral nerve injury. Neuroscience. 2010;166:1–4. doi: 10.1016/j.neuroscience.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 61.van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155:654–662. doi: 10.1016/j.pain.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 62.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woolf CJ, Mannion RJ. Neuropathic pain: aetiology, symptoms, mechanisms, and management. Lancet. 1999;353:1959–1964. doi: 10.1016/S0140-6736(99)01307-0. [DOI] [PubMed] [Google Scholar]
- 64.Woolf CJ, Salter MW. Neuronal plasticity: increasing the gain in pain. Science. 2000;288:1765–1769. doi: 10.1126/science.288.5472.1765. [DOI] [PubMed] [Google Scholar]
- 65.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan S, Shi Y, Tang SJ. Wnt signaling in the pathogenesis of multiple sclerosis-associated chronic pain. J Neuroimmune Pharmacol. 2012;7:904–913. doi: 10.1007/s11481-012-9370-3. [DOI] [PubMed] [Google Scholar]
- 67.Zhang YK, Huang ZJ, Liu S, Liu YP, Song AA, Song XJ. WNT signaling underlies the pathogenesis of neuropathic pain in rodents. J Clin Invest. 2013;123:2268–2286. doi: 10.1172/JCI65364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Z, Cai YQ, Zou F, Bie B, Pan ZZ. Epigenetic suppression of GAD65 expression mediates persistent pain. Nat Med. 2011;17:1448–1455. doi: 10.1038/nm.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhuo M. Cortical excitation and chronic pain. Trends Neurosci. 2008;31:199–207. doi: 10.1016/j.tins.2008.01.003. [DOI] [PubMed] [Google Scholar]