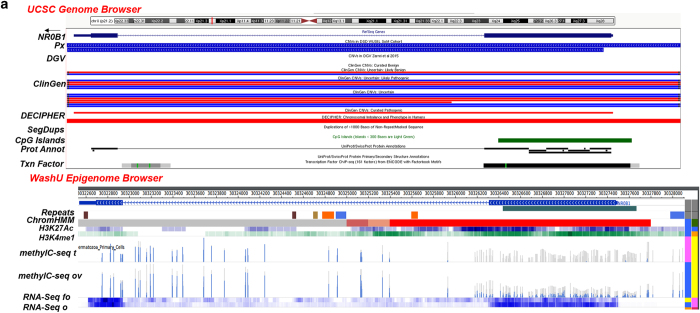
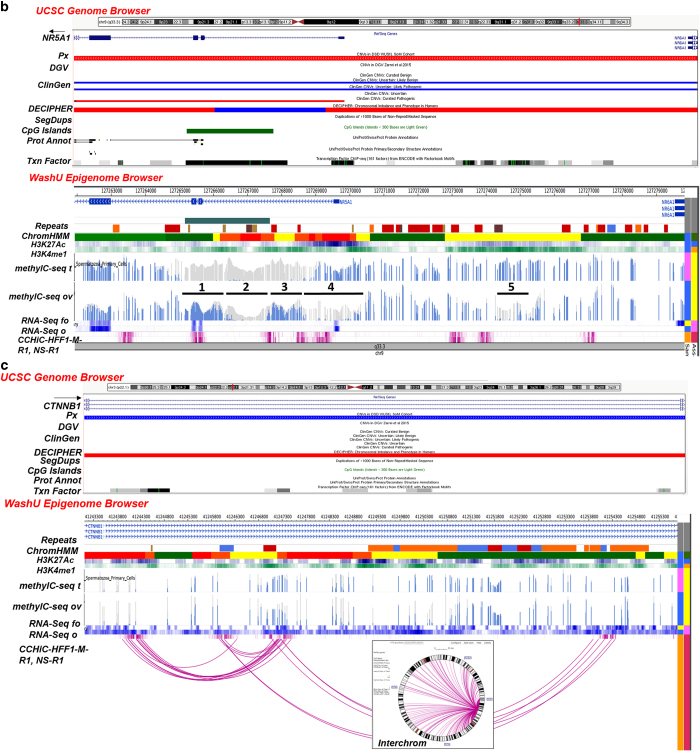
Figure 1.
(a) CN gains overlapping with an intact NR0B1 (5 kb; Xp21.2). UCSC Browser—CN gains (blue) overlapping with an intact NR0B1 (exons 1–2) in 2 patients (Px); D28 (XY) with underdeveloped genitalia (5.6 Kb, 39 markers; chrX:30,322,475–30,328,115) and D30 (XXY) with ambiguous genitalia, undescended testes and small ureters (4.9 kb, 32 markers; chrX:30,322,476–30,327,408). DGV (Zarrei et al.,13)—no overlapping CNVs found. ClinGen—CN gains and losses categorized as uncertain clinical significance (UCS), UCS likely benign (UCS LB), UCS likely pathogenic (UC LP) and a pathogenic loss. DECIPHER—losses in patients with variable findings. Segmental Dups (SegDups)—none. CpG Islands—exon 1. Protein Annotation (Prot Annot)—ligand binding, AF-2 motif, LXXLL motifs 1–3, AA tandem repeat and repeats 1–4. Transcription Factor (Txn Factor)—exons 1 and 2. WashU Epigenome Browser. Repeat Masker (Repeats)—intron 1: LINE (orange), DNA transposon (blue), simple repeat, microsatellite (SR/MS; gold), satellite repeat (SR; brown). ChromHMM—active transcription start site (aTSS) (red); repressed polycomb (gray). Histone Modification—H3K27Ac (acetylation) and H3K4me1 (methylation) marks in the ovary. DNA Methylation (methylC-seq t, ov)—differential profile in exon 1—some DNA in fetal ovary and none in fetal testis). RNA Expression (RNA-Seq fo, o)—differential profile—more in adult ovary than in fetal ovary. (b) CN loss overlapping with partial region of NR5A1 (26 kb; 9q33.3). UCSC Browser—CN loss (red) involving exon 1-intron 4 in a patient (Px D9; XY) with hypospadias (18 kb; 5 markers; chr9:127,261,935–127,279,829). DGV (Zarrei et al.,13)—no entry. ClinGen—UCS likely benign (LB) gain, pathogenic loss, UCS LP gain. DECIPHER—loss/gain (Note: truncated exon 1 of NR6A1 at the 3' end of the CNV). Segmental Dups (SegDups)—none. CpG Islands—intron 1-exon 2-intron 3. Protein Annotation (Prot Annot)—dimerization, ligand binding, phosphorylation, cross-linking, DNA binding, Zn finger, acetylation. Transcription Factor (Txn Factor)—exons 1-intron 4, upstream of NR5A1. WashU Epigenome Browser. Repeat Masker (Repeats)—exon 1-intron 4—SINEs (red), LINE (orange), LCRs (dark brown), SR/MS (gold); upstream of NR5A1—SINEs (red), LINE (orange), LCRs (dark brown), SR/MS (gold), DNA transposon (blue). ChromHMM—exon 1-intron 4, active transcription start site (aTSS) (red); enhancers (yellow), strong transcription site (green), weak TS (dark green); upstream of NR5A1—aTSS, enhancers, weak TS. Histone Modification—some H3K4me1 methylation and H3K27Ac acetylation marks in ovary. DNA Methylation (methylC-seq t, ov) in 5 regions involving NR5A1 (numbered and underlined)—absence of DNA methylation in fetal testes in exons 2–3 (1) and intron 1 (3) overlapping with an enhancer; absence of methylation in intron 1 overlapping with an aTSS in both fetal testis and ovary (2); absence of methylation in ovary in exon 1 (3) and upstream of NR5A1 overlapping with an enhancer (5). RNA Expression (RNA-Seq fo, o)—similar RNA expression in adult and fetal ovaries. Local Interactions (CCHiC-HFF1-M-R1/NS-R1)—various regions within and upstream of NR5A1 are locally interacting (purple). (c) CN gain involving intron 1 of CTNNB1 (41 kb, 3p22.1). UCSC Browser—CN gain (blue) overlapping with intron 1 of CTNNB1 in a patient (Px D14, XY) with ambiguous genitalia (12.6 kb, 59 markers; chr3:41243113–41255684). DGV (Zarrei et al.,13)—no overlapping CNVs found. ClinGen—no overlapping CNVs found. DECIPHER—losses in patients with variable findings. Segmental Dups (SegDups)—none. CpG Islands—none. Protein Annotation (Prot Annot)—none. Transcription Factor (Txn Factor)—several. WashU Epigenome Browser. Repeat Masker (Repeats)—LINEs (orange), SINEs (red), DNA transposons (blue). ChromHMM—active transcription start site (aTSS) (red); enhancers (yellow), weak TS (dark green). Histone Modification—some H3K4me1 methylation and H3K27Ac acetylation marks in ovary. DNA Methylation (methylC-seq t, ov)—differential DNA methylation in fetal testes and fetal ovaries. RNA Expression (RNA-Seq fo, o)—differential profile in adult and fetal ovaries. Local Interactions (CCHiC-HFF1-M-R1/NS-R1)—various regions within and upstream of NR5A1 are locally interacting (purple arcs). Interchromosomal Interactions (Interchrom inset)—interactions of intron 1 of CTNNB1 with other chromosome loci (purple lines; details in Supplementary Figure 2A). CNV, copy number variation; LCRs, low copy repeats; LINEs, long interspersed elements; SINEs, short interspersed elements.