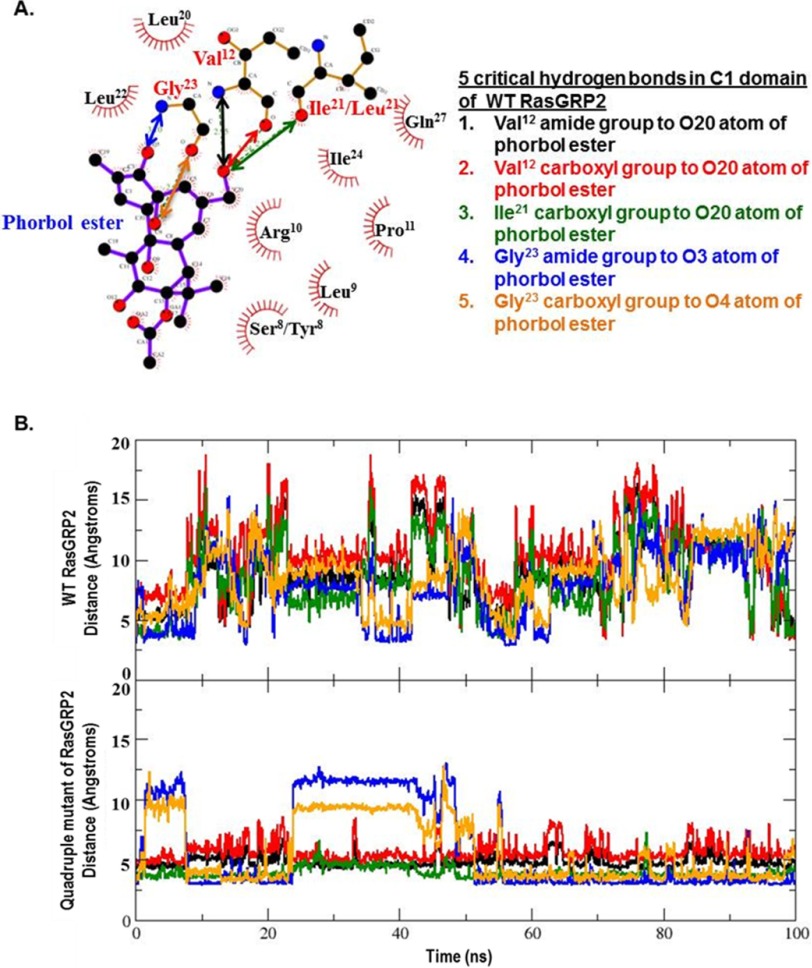
FIGURE 10.
Interactions between the critical binding residues in the phorbol ester and the C1 domain. A, the distances for the critical binding interactions between phorbol ester and the C1 domain, as determined from the crystallographic structure of the binary binding complex with the C1b domain of PKCδ, were evaluated for the wild-type and quadruple mutant of RasGRP2. Ser/Tyr8 and Ile/Leu21 indicate the corresponding residues in the wild-type and mutant, respectively. The image is from LIGPLOT taken from the website PDBsum (54). Oxygen is red, nitrogen is blue, and carbon is black. The covalent bonds in the protein are in brown; those in the phorbol ester are in purple. Amino acid numbering is based on the position in the C1 domain or, in brackets, the position of the amino acid in PKCδ. B, distances calculated for the centers of mass of the interacting partners over the interval of the molecular dynamics simulations for the wild-type and quadruple mutant C1 domain of RasGRP2 as a function of stimulation time. Colors are for the interacting pairs as indicated in A.