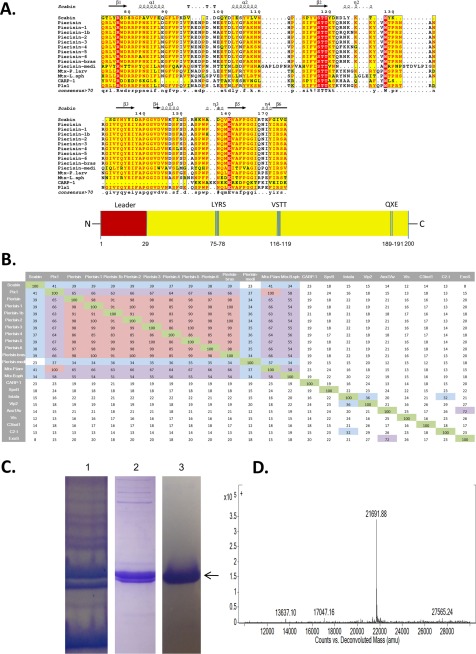
FIGURE 1.
Multiple-sequence alignment of Scabin with various Pierisin-like mART toxins. A, sequence alignment of Scabin with some Pierisin-like toxins produced using the T-Coffee Web server to align the sequences and ESPript to generate the alignment figure (45). Key catalytic regions are highlighted. Identical residues are highlighted in red, and similar residues are printed in red type. B, identity matrix showing the amino acid identity between the 100 core catalytic residues of the known ExoS-like, C2-like toxins and Vis. Salmon, highly diverse sequences; light green, a large amount of conservation; yellow, an intermediate level of conservation between sequences. The identity matrix was generated using ClustalX2 (33) and colored using Microsoft Excel. C, purification and identification of Scabin from E. coli lysate. SDS-polyacrylamide gels showing the protein banding pattern for crude lysate (lane 1), immobilized metal affinity chromatography purification (lane 2), and FPLC ion exchange chromatography (lane 3). The arrow indicates the position of the Scabin protein. D, Q-TOF mass analysis of purified Scabin protein showing a single peak at 21,691.9 Da, corresponding to the expected mass of recombinant Scabin.