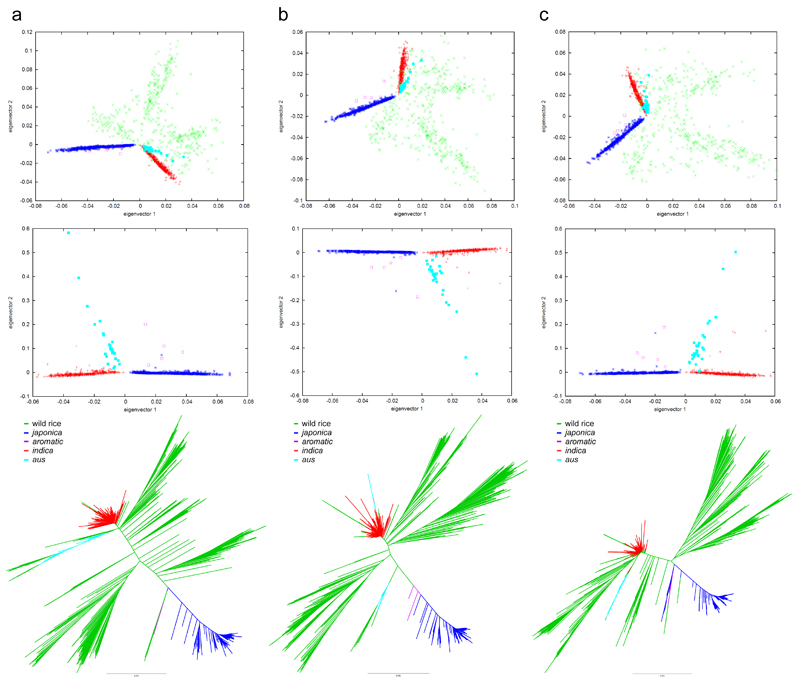
Figure 2.
Population structure analysis of low-variation regions in cultivated rice genomes. a, CLDGR threshold 3; b, CLDGR threshold 4; c, CLDGR threshold 5. Top panels, PCAs for all accessions; middle panels, PCAs without wild populations; bottom panels, unrooted NJ trees of concatenated CLDGRs. All shown eigenvectors discriminate the domesticated groups with statistical significance except: the first eigenvector fails to discriminate aromatic and japonica in all cases; the second eigenvector fails to discriminate aromatic and aus in all cases. Consequently, the aromatic accessions occupy the space between aus and japonica, indicating that aromatic could be an aus×japonica hybrid.