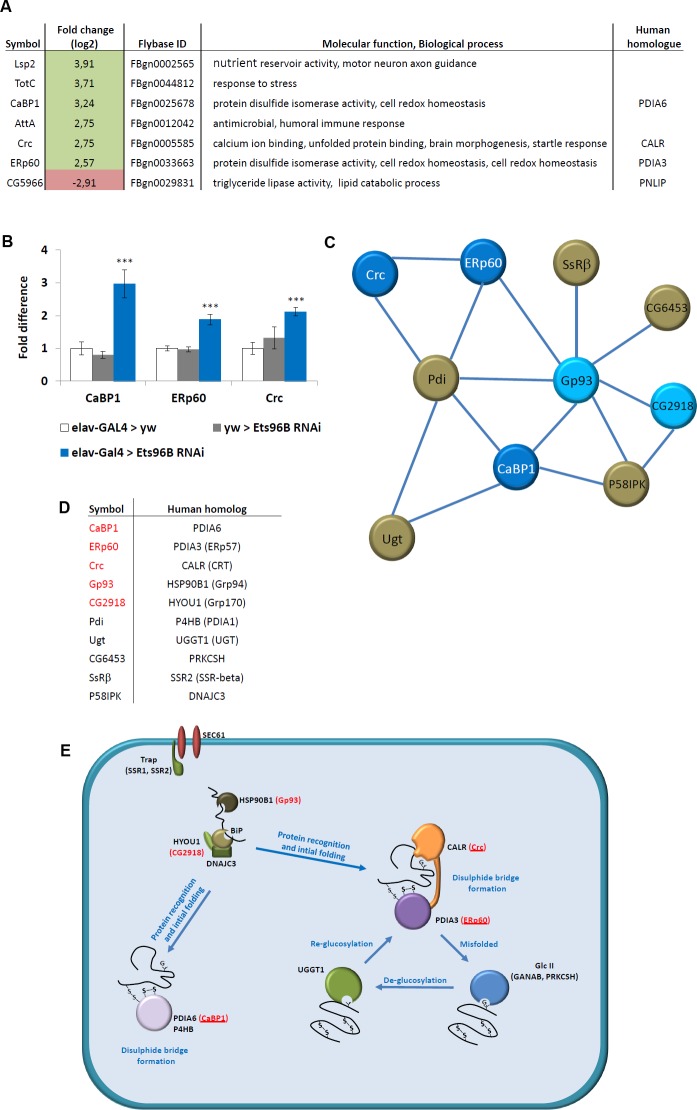
Fig 3. Ets96B regulates a conserved molecular chaperone complex.
(A) SOLiD data for elav-GAL4>Ets96BRNAi versus controls (w, elav-GAL4>w1118 and w1118> Ets96BRNAi) 5–7 day old male heads. Only genes regulated more than ±log2 2.5-fold change are included. (B) Relative expression levels of CaBP1, ERp60 and Crc in 5–7 day old control males or males where Ets96B was knocked down in the entire nervous system throughout development. This assay was repeated at least 7 times. Error bars indicate SEM. (n = 25 males per treatment; *** P<0.005 compared with elav-GAL4 controls, one-way ANOVA with Tukey’s post hoc test for multiple comparisons). (C) Biogrid and String were used to find all genes that interact with the molecular chaperone genes recovered in the Ets96B knockdown males. Genes in red were recovered in the Ets96B SOLiD sequencing. (D) Data showing human homologues of Drosophila genes recovered in Biogrid and String analysis. Genes in red were recovered in the Ets96B SOLiD sequencing. (E) Diagram depicting molecules involved in endoplasmic reticulum protein folding. These include molecular chaperones that function to prevent aggregation (Bip, HSP90B1, DNAJC3, HYOU1 and CALR), protein disulfide isomerases that catalyze disulfide bond formation, isomerization and reduction (PDIA3, PDIA3 and P4HB), and proteins involved in de- or re-glycosylation of improperly folded glycoproteins (PRKCSH and UGGT1). Drosophila homologues that were up-regulated when Ets96B was knocked down are in red, and those that were up-regulated more than 2.5-fold are underlined.