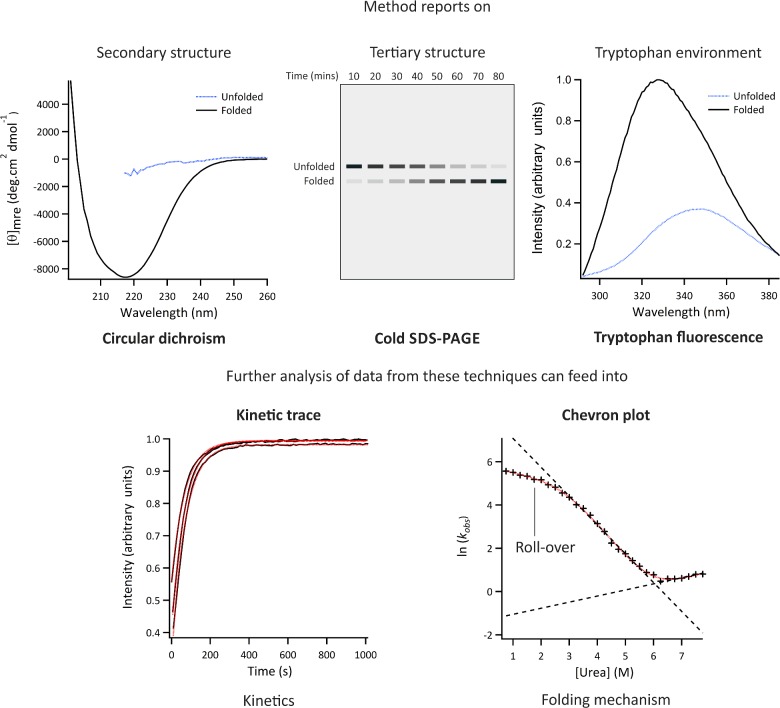
Figure 3. ‘Classic’ methods of interrogating protein folding.
Different techniques provide independent, and complementary, information about the kinetics, thermodynamics and mechanism of folding. These approaches have been used for analysis of water soluble and OMP folding (see text). CD reports on the difference in absorbance of left and right circularly polarized light by peptide bonds in an asymmetric environment. In this case, the asymmetric environment refers to the protein's secondary structure, with e.g. β-sheet, α-helix and disorder each giving rise to characteristic spectra. CD in the far UV can be used for the analysis of both water soluble proteins and OMPs. [θ]mre; mean residue ellipticity. Cold SDS-PAGE is useful for the analysis of the formation of native OMPs since these structures are resistant to denaturation in SDS without heating. This method involves initiating a folding reaction, taking samples at particular time points and quenching further folding with SDS, then running the samples on an SDS-PAGE gel without boiling. These gels can be analysed qualitatively or quantitatively by measuring the band intensity through densitometry. Tryptophan fluorescence is sensitive to the polarity of the local environment. Upon folding, tryptophan residues which move to more hydrophobic environments (such as within a folded protein core, or interfaced with lipid membranes or detergents) show a characteristic ‘red-shift’ in their emission maxima and, usually, an increase in fluorescence intensity. Kinetic traces can be obtained by monitoring the change in signal intensity from CD and/or tryptophan fluorescence at particular wavelengths or, for OMPs, by analysing samples taken at different refolding times using cold SDS-PAGE. Chevron plots involve measuring the rate constants for folding/unfolding from kinetic traces measured in increasing concentrations of denaturant and plotting the natural logarithm of these values as a function of denaturant concentration. Red lines indicate calculated fits.