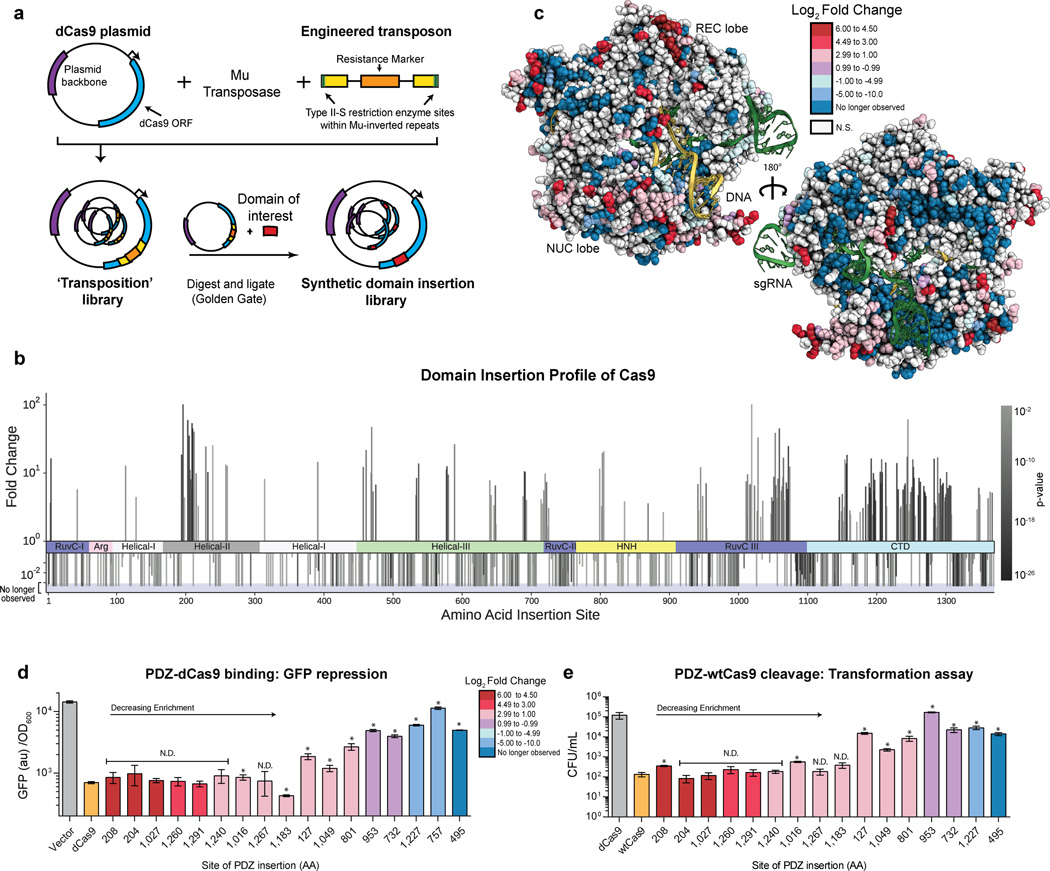
Figure 1. Mapping the insertion potential of Cas9 with the Alpha-Syntrophin PDZ protein interaction domain.
(a) Generation of the transposon-based domain insertion library. (b) Fold change values for insertions at specific amino acid sites derived from sequencing data over two rounds of screening. A positive value indicates the preference of the domain insertion at a site to remain in the library after screening for function. A negative value indicate a loss of the clone with an insertion at the site. More significant P values (DESeq, multiple hypothesis testing corrected) are represented as darker color bars. Positive values which attain 102 represent sites that were not sequenced before screening. Negative bars that extend into the shaded region represent clones which have been cleared from the library (i.e. not observed after screening). (c) Log2 fold change values from (b) mapped onto the structure of Cas9 (PDB ID:4UN3)11. (d) GFP repression activity of individual PDZ insertion sites. Values represent biological replicates with standard deviation (n=3), constructs are in order of decreasing fold change from sequencing. Positive and negative controls dCas9 and vector only are colored orange and grey, respectively. N.D. stands for no difference detected from dCas9, t-test (p > 0.01), (* = p < 0.01) (e) Cleavage activity of clones via an E. coli based transformation assay. Cas9 activity results in genomic cleavage and lower CFU/mL, values represent biological triplicates with standard deviation (n=3). Positive and negative controls Cas9 and dCas9 are colored orange and grey, respectively. N.D. stands for no difference detected from WT Cas9, t-test (p > 0.01), (* = p < 0.01)