Fig. 6.
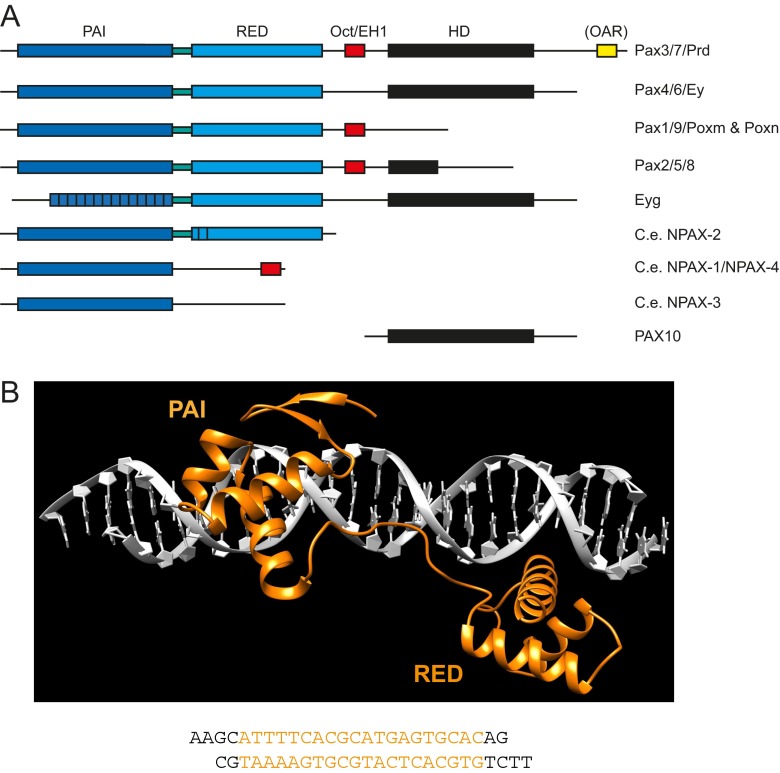
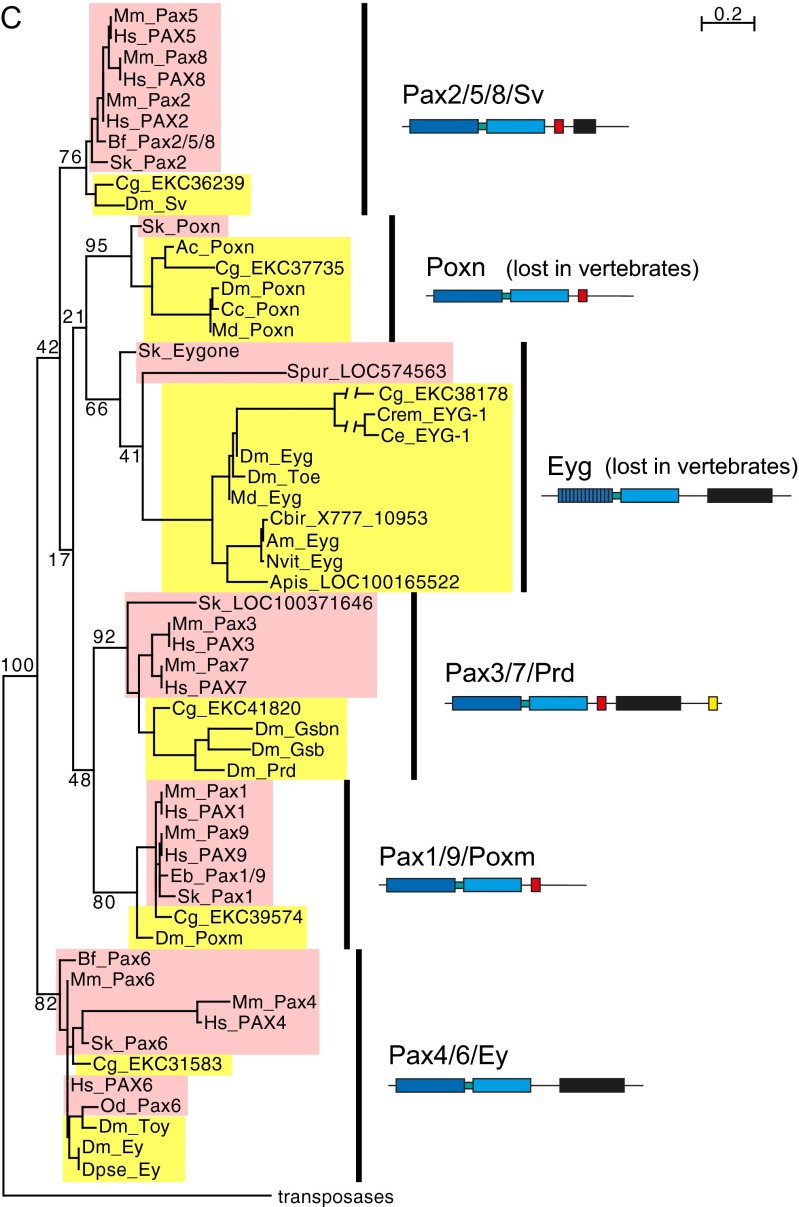
PRD class of homeobox genes. a Schematic view of the variable domain and motif organization found in different PRD families or proteins. The PAIRED domain is composed of two subdomains, PAI and RED, separated by a linker. Brackets indicate motifs not present in all genes of a family. b Structure of the PAIRED domain of human PAX6 bound to DNA (PDB ID: 6PAX) (Xu et al. 1999). PAI and RED domains are indicated. The bound sequence is shown underneath. The beta-strands and alpha helices are marked in the sequence alignment in Sup. Fig. S5. c Rooted phylogenetic tree of bilaterian PRD class proteins, based on the PAIRED domain. The PAIRED domain similarity region of four bilaterian transposases (from acorn worm and oyster) was used as outgroup. Values of 100 bootstraps values are shown at selected clades. On the right side, the PRD class families are indicated, together with their typical structural organization. Deuterostome branches are highlighted in pink, protostome branches in yellow. Branch lengths in the Eyg family were reduced where indicated (Ce, Crem, CG). Note that the branching of the six families should not be taken as evidence for how they evolved from an ancestral precursor. Drosophila proteins: Sv: Shaven; Poxn: Pox-neuro; Poxm: Pox-meso. The phylogenetic tree was created using PhyML as implemented in SeaView (Gouy et al. 2010). About 100 residues containing the C-terminal region of PAI and the complete RED subdomain from the multiple sequence alignment in Sup. Fig. S5 were used for tree generation. For species codes, see Sup. Fig. S5