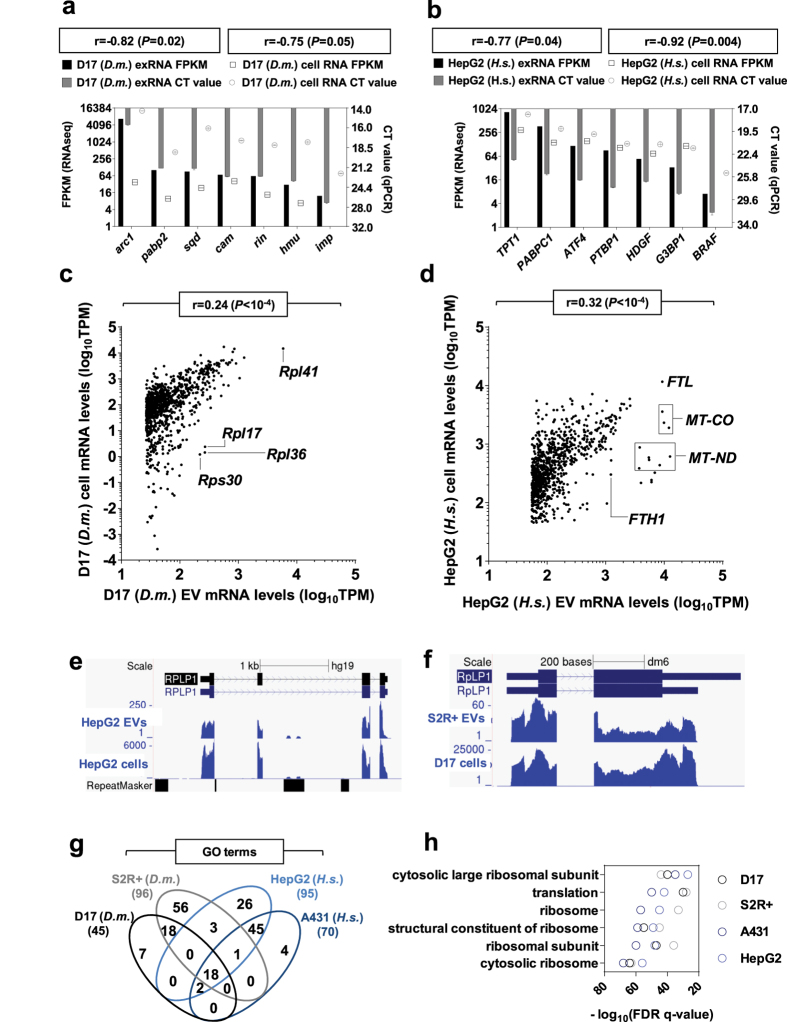
Figure 5. Characterization of mRNAs secreted within Human and Drosophila EVs.
(a,b) Comparative analysis of expression levels of select mRNAs via RNA-seq and qRT-PCR. Cycle threshold (CT) values determined by qRT-PCR are negatively correlated to FPKM values determined by RNA-seq for various Drosophila D17 (a) and human HepG2 (b) mRNAs in EVs and cells. (c,d) Relative expression levels of mRNAs extracted from D17 (c) or HepG2 cells and EVs (d). Select groups of transcripts are identified. Pearson’s correlations (r) and associated p-values are indicated at the top of each graph. (e,f) UCSC genome browser views of Rplp1 mRNA shows strictly exonic read coverage in human HepG2 (e) and Drosophila D17 (f) EVs and cells. (g) Venn diagram depicting the overlap of enriched gene ontology (GO) terms displayed for human and Drosophila EVs mRNAs. The number of enriched GO terms retrieved per sample is shown. (h) Examples of translation-related GO terms identified in all EV samples. Associated false discovery rates (FDR) are provided.