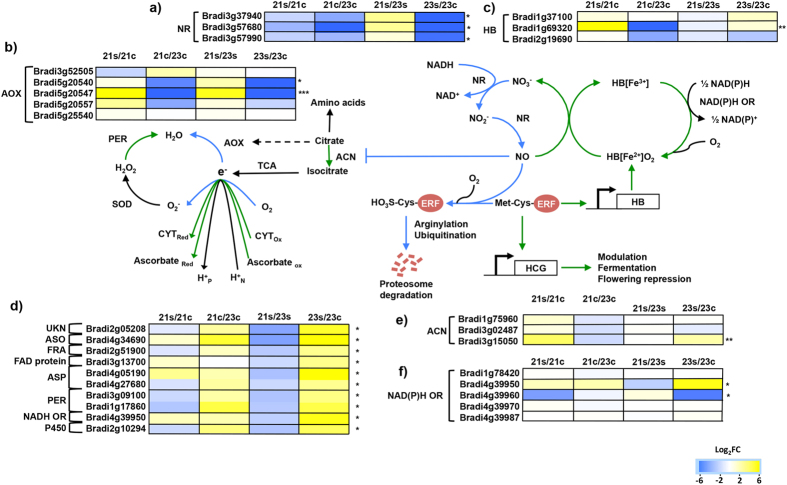
Figure 3. Integration of differentially expressed transcripts related to ROS management in tolerant and sensitive ecotypes of Brachypodium distachyon under submergence stress.
(a) NITRITE REDUCTASE (NR). (b) ALTERNATIVE OXIDASE (AOX). (c) HAEMOGLOBIN (HB). (d) Transcripts involved in different ROS end-detoxification pathways: UKN (unknown), ASO (ASPARTATE OXIDASE), FRA (FRATAXIN), ASP (ASCORBATE PEROXIDASE), PER (PEROXIDASE). (e) ACONITASE (ACN). (f) NAD(P)H oxidoreductase (OR). Blue indicates down-regulation and yellow indicates up-regulation in Log2FC values after 48 h stress measured by RNA-Seq. The letters s and c indicate stress and controls, and the numbers 21 and 23 indicate Bd21 and Bd2-3, respectively. The green and blue arrows indicate up- and down-regulated activity in tolerant Bd2-3. *Significantly up- or down-regulated in Bd2-3,**significantly up- or down-regulated in Bd21,***significantly and inversely regulated in Bd2-3 and Bd21 (FDR < 0.05 × 10−5; Log2FC ≥ 1.5 or ≤ −1.5). Acronyms: SOD (superoxide dismutase), HCG (hypoxia core genes), TCA (tricarboxylic acid cycle), ERF (ethylene responsive factor), and NO (nitric oxide).