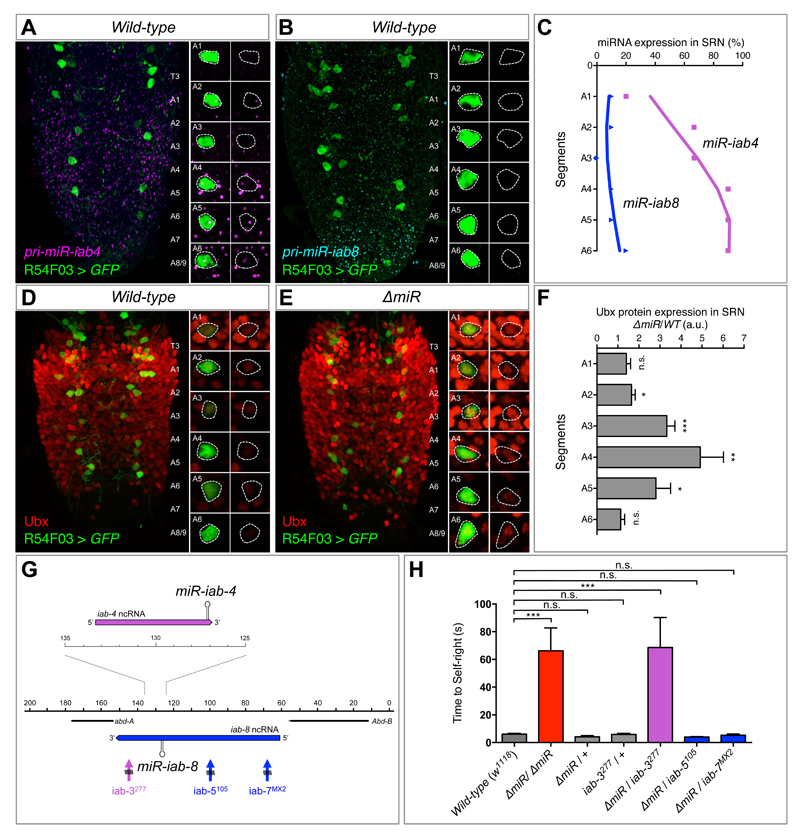
Figure 3. Regulation of Ubx protein expression in SRN cells by miR-iab4/iab8.
(A) Wild type expression of precursor miR-iab4 transcripts (RNA-FISH, magenta) in SRN cells (R54F03>GFP, green) of the ventral nerve cord (VNC) of first-instar Drosophila larvae. (B) Wild type expression of precursor miR-iab8 transcripts (RNA-FISH, cyan) in SRN cells (R54F03>GFP, green) of the VNC of first-instar Drosophila larvae. (C) Percentage of SRN cells expressing miR-iab4 (purple, square) and miR-iab8 (blue, triangle) precursors across A1 to A6 (N = 10). (D and E) Ubx protein expression (red) in SRN cells of wild type (D) and ∆miR (E) first-instar larvae VNCs. (F) Quantification of Ubx protein expression ratio of ∆miR over wild type within the SRN cells (red) by fluorescent intensity (N = 8 per genotype; arbitrary units, a.u.). (G) Diagram of a sub-region of the bithorax complex based on (13) showing iab-4 (purple) and iab-8 (blue) non-coding RNAs (ncRNA), and rearrangement breakpoints affecting miR-iab-4 (iab-3277, purple) and miR-iab-8 (iab-5105 and iab-7MX2, blue). (H) Genetic complementation tests to determine the involvement of miR-iab4 or miR-iab8 in SR behaviour using trans-heterozygote larvae for ∆miR and different chromosomal rearrangement breakpoints that disrupt the bithorax complex (mean ± SEM; N = 17 to 20 per genotype). A non-parametric Mann-Whitney U test was performed to compare treatments; p>0.05 (non-significant; n.s.); p < 0.05 (*); p < 0.01 (**); p<0.001 (***).