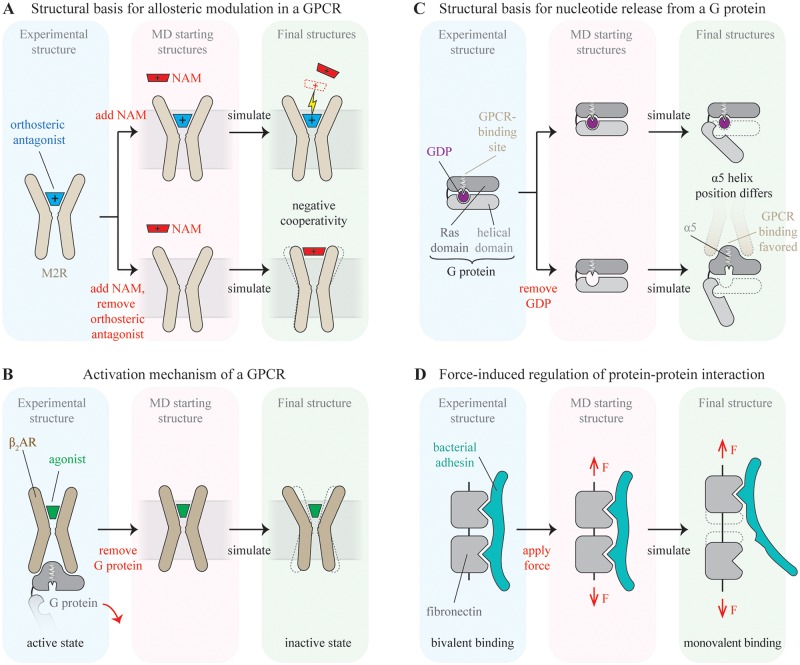
Fig 2. Simulation schematics for case studies.
Key perturbations to the experimental structure are indicated in red text. (A) Binding and cooperativity of a negative allosteric modulator (NAM) in the M2 muscarinic receptor (M2R). The binding pose of the NAM and its cooperativity with the orthosteric ligand are probed by performing unguided binding simulations with an unliganded receptor and a receptor with bound orthosteric ligand. Negative cooperativity leads to stronger binding of the NAM to the unliganded M2R. Simulations indicate that cooperativity in this system is due both to direct electrostatic repulsion between cationic ligands and coupled conformational changes of the two binding sites. (B) Activation mechanism of the β2-adrenergic receptor (β2AR). Removal of the bound G protein from the active-state, agonist-bound crystal structure of β2AR leads to a spontaneous transition to the inactive state in simulation, capturing the activation process in reverse. (C) Mechanism of GPCR-catalyzed nucleotide release from a heterotrimeric G protein. Simulations of a G protein with and without bound guanosine diphosphate (GDP) suggested that nucleotide release from a G protein—which leads to G protein activation—takes place via a previously unexpected mechanism. (D) Force-induced uncoupling of a bacterial adhesin from a fibronectin fragment. MD simulations of a fibronectin–adhesin complex led to the discovery that application of stretching forces to fibronectin reduces its affinity to adhesin.