Fig.1.
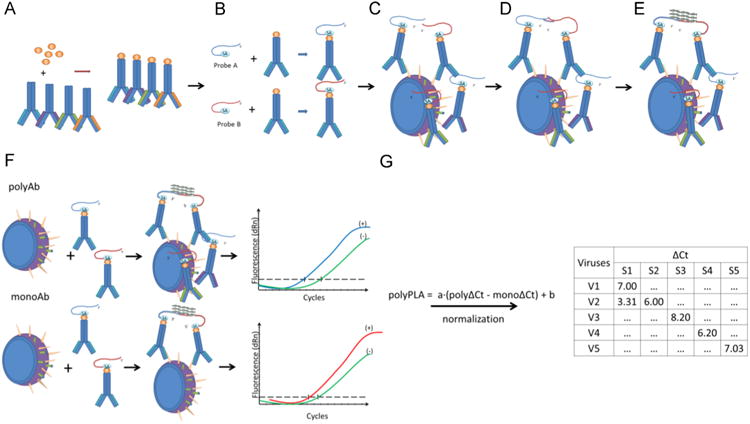
The simplified diagram of polyPLA. PolyPLA quantifies the antibody antigen binding avidity using the amplification signals in quantitative PCR (qPCR) from the pairs of primers attached to a reference polyclonal antiserum. First, polyPLA biotinylates a reference polyclonal antiserum (A), which will be then labeled with sodium azide-linked 5′ and 3′ oligonucleotides (B). A labeled polyclonal antiserum with ΔCt≥8.5 in the ligation efficiency test is then incubated with a reference antigen (virus) or a testing antigen (C), followed by the approximate ligation of the two oligos (D). The antibody antigen binding avidity is quantified using the amplification signals ΔCt in qPCR (E). The ΔCt values among the polyclonal antisera and antigens can be compared to assess antigenic differences among these tested antigens, and these ΔCt values can be viewed as similar as the serological titers, such as HI and neutralization titers, from conventional serological assays (F). The polyPLA units were normalized by its ΔCt values for polyclonal antiserum (polyΔCt) with its ΔCt values for monoclonal antibody against NP (monoΔCt) (G).
