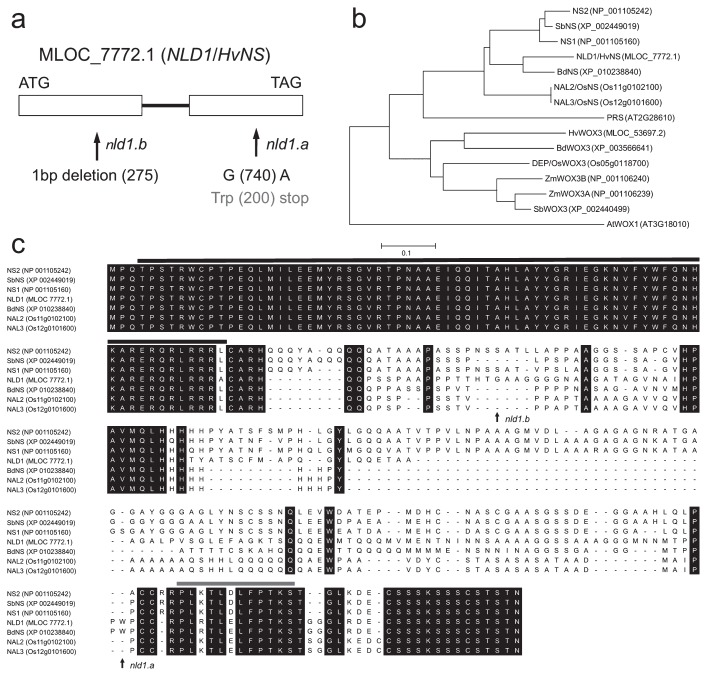
Fig. 6.
Molecular characterization of the NLD1 gene. (a) Exon-intron structure and the mutation sites of nld1.a and nld1.b. Exons and intron are shown by boxes and bold line, respectively. (b) Phylogenetic tree of NLD1-related proteins in barley (NLD1; MLOC_7772.1, HvWOX3; MLOC_53697.2), maize (NS1; NP_001105160, NS2; NP_001105242, ZmWOX3A; NP_001106239, ZmWOX3B; NP_001106240), rice (NAL2; Os11g0102100, NAL3; Os12g0101600, DEP; Os05g0118700), sorghum (SbNS; XP_002449019, SbWOX3; XP_002440499), Brachypodium distachyon (BdNS; XP_010238840, BdWOX3; XP_003566641), and Arabidopsis thaliana (PRS; AT2G28610, AtWOX1; AT3G18010). The tree was created using MEGA ver. 5.2.2 (available at http://megasoftware.net/, Tamura et al. 2011). (c) Comparison of the amino acid sequences among NLD1-related proteins in the NS cluster. Amino acid sequences were aligned with MUSCLE program of MEGA ver.5.2.2. Amino acids conserved in all proteins are represented by black boxes. Mutation sites in nld1.a and nld1.b are indicated with arrows. Black line and gray line indicate the homeobox domain and the WUS box motif, respectively.