Figure 6.
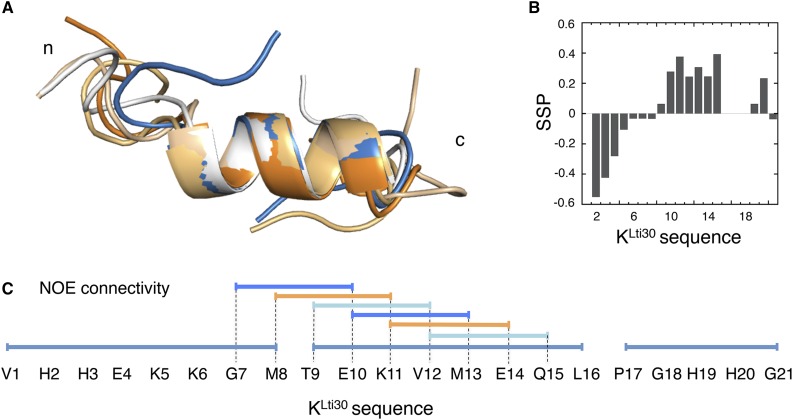
Structural analysis of membrane-bound KLti30 peptide by NMR. A, The five best structures of bicelle-bound KLti30 peptide from 1H-NMR constraints, where the central nine residues of the peptide adopt a fixed α-helix, whereas the N and C termini are more disordered. B, Secondary structure propensity (SSP) from 1Ha, 1HN, and 1Hb chemical shifts, where positive values show the induced α-helix in the central part of the peptide. Negative secondary structure propensity values indicate extended conformation in the N-terminal region of the peptide. C, Determined NOE connectivity between peptide residues along the KLti30. The ordered n+3 couplings within the residue segment Gly-7 to Gln-15 are the hallmark of the α-helical structure.