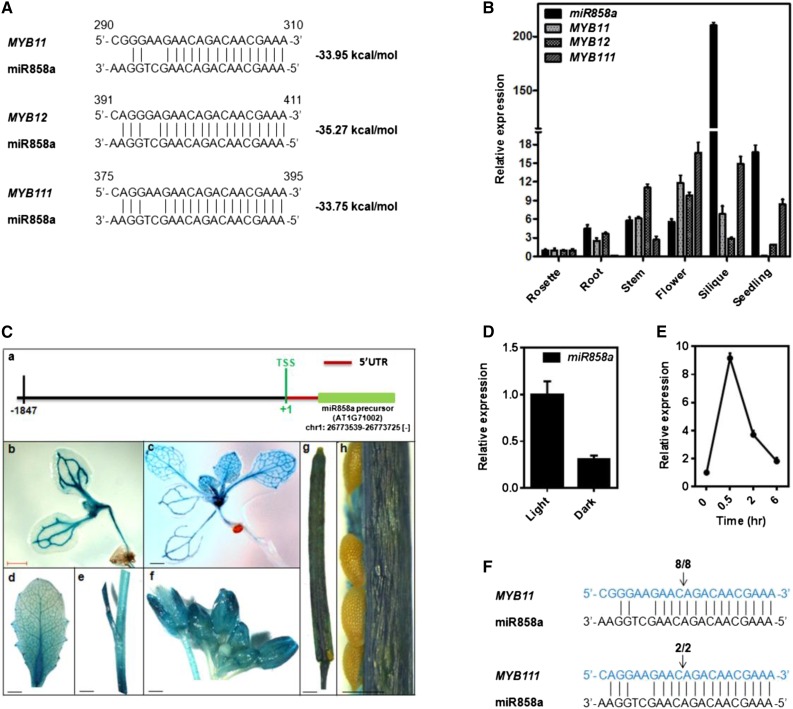
Figure 1.
Identification and characterization of miR858a in Arabidopsis. A, Sequences of MYB11, MYB12, and MYB111 mRNAs (5′ to 3′) aligned with the mature sequence of miR858a (3′ to 5′). Dashed lines and spaces represent perfect match and G:U wobble pairing, respectively. The free energy of binding is indicated at right. B, Expression of miR858a, MYB11, MYB12, and MYB111 in rosette, root, stem, flower, and silique from 5-week-old plant and 10-d-old seedling of Arabidopsis Columbia-0 (Col-0). For miR858a, SnoR41Y RNA was used, and for MYB11, MYB12, and MYB111, TUBULIN was used as the endogenous control to normalize the relative expression levels. Bars represent means ± se (n = 3). C, Histochemical staining showing GUS activity in different developmental organs in the ProMIR858:GUS transgenic lines: a, schematic representation of the miR858a promoter upstream of the precursor; b, 5-d-old seedling; c, 12-d-old seedling; d, rosette leaf; e, stem; f, inflorescence; g, silique; and h, seed. UTR, Untranslated region. Bars = 1 cm for b to f and 500 µm for g. D, qRT-PCR analysis of the mRNA level of miR858a in wild-type (Col-0) seedlings grown in light and dark conditions for 5 d. Bars represent means ± se (n = 3) from three biological replicates. E, qRT-PCR analysis of the mRNA levels of miR858a in wild-type upon exposure to light (150 µmol m−2 s−1) at the indicated times. Bars represent means ± se (n = 3) from three biological replicates. F, Target validation for miR858a by RLM-RACE. Cleavage products of MYB11 and MYB111 mRNA are shown. The fractions of clones with the expected 5′ end among all sequenced products are indicated above the target site. Arrows indicate the cleavage sites detected by RLM-RACE.