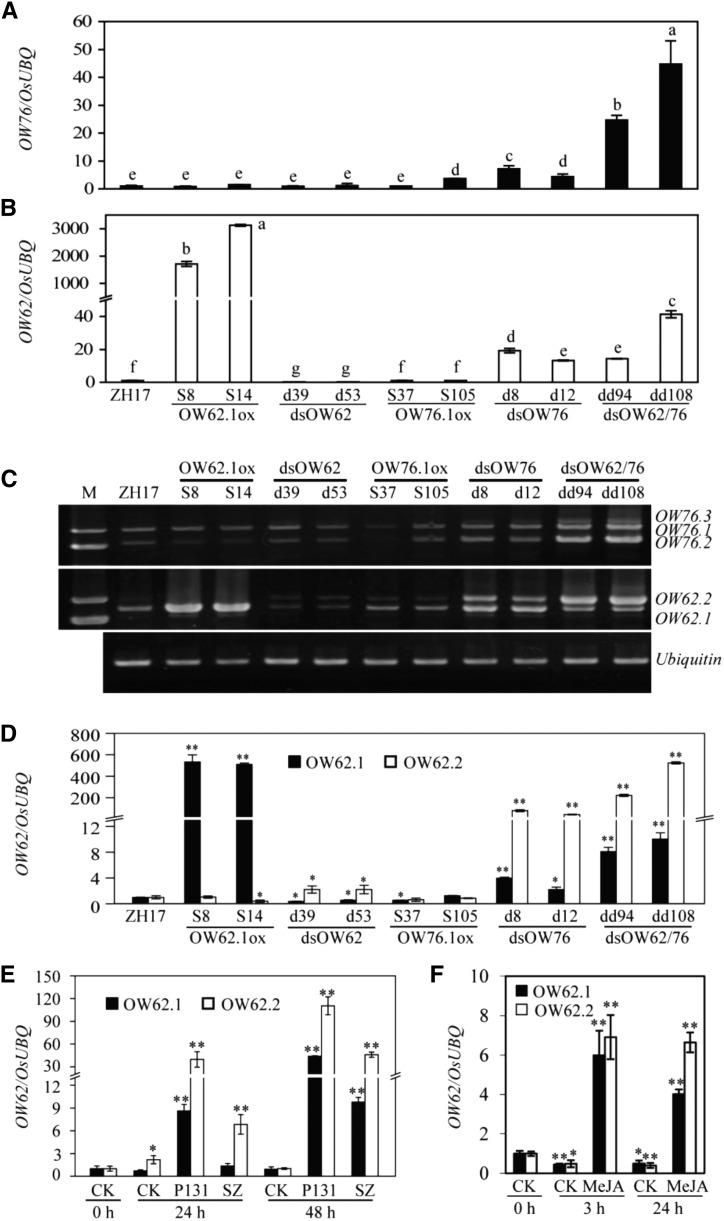
Figure 6.
Changes in alternative transcript levels of OsWRKY62 and OsWRKY76 in the RNAi plants or plants after pathogen infection or MeJA treatment. A and B, Expression of OsWRKY62 and OsWRKY76 was analyzed by qRT-PCR using total RNA extracted from leaves of 3-week-old plants. Error bars indicate se (n = 3). Values marked with different letters indicate statistically significant differences as analyzed by the SAS software (Duncan’s multiple range test, α = 0.05). C, OsWRKY76.1 to OsWRKY76.3 (OW76.1–OW76.3) and OsWRKY62.1 and OsWRKY62.2 (OW62.1 and OW62.2) were obtained by RT-PCR amplification of complementary DNAs (cDNAs) from dd94 and dd108 plants. D to F, Levels of alternative transcripts OsWRKY62.1 (OW62.1) and OsWRKY62.2 (OW62.2) were analyzed by qRT-PCR in transgenic plants (D), after M. oryzae infection (E), or after MeJA treatment (F). Samples were collected from plants at the time points indicated after inoculation with a virulent (SZ) or avirulent (P131) strain of M. oryzae or after treatment with MeJA (0.1 mM, in 10 mM MES buffer, pH 6). CK, Mock treatment. Transcription levels are shown relative to the level of transcript at 0 h or those in wild-type ZH17 using the OsUBQ gene as an internal standard. Values shown are means ± se of three replicates. Statistically significance changes are indicated by asterisks: *, P < 0.05 and **, P < 0.01 (Student’s t test).