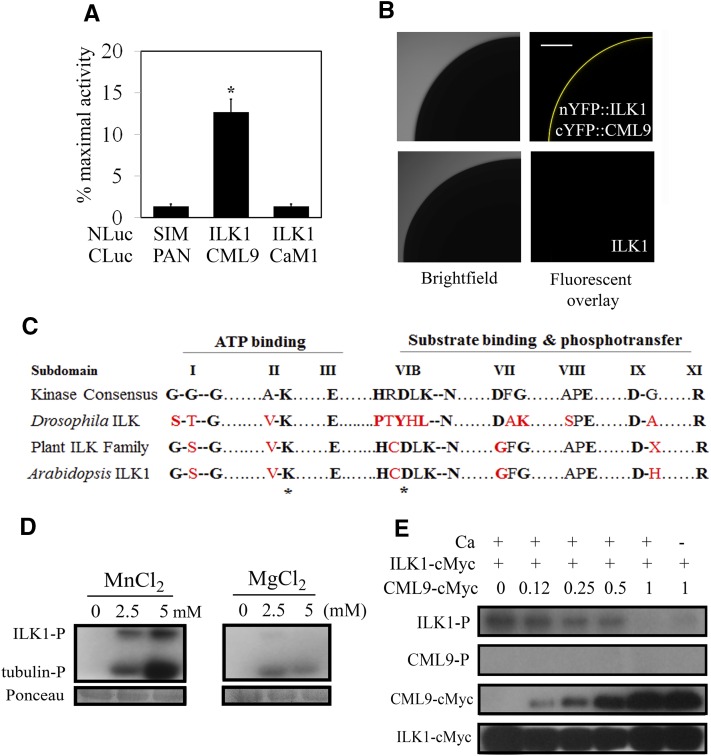
Figure 1.
ILK1 interacts with CML9 in vivo and is an active kinase in vitro. A, The SLCA in Arabidopsis protoplasts revealed interactions between the bait (NLuc-ILK1) and prey (CLuc-CML9) fusion proteins by quantifying the amount of luciferase activity relative to that of the HISTONE 2A and 2B interactors (NLuc-H2A, CLuc-H2B; Fujikawa and Kato, 2007). The response of the negative control (NLuc-SIM, CLuc-PAN) is shown for reference. Data from four replicates from three independent experiments for each pair are shown where * indicates significant difference from the control, P < 0.05). B, Images showing interaction of ILK1 with CML9 by bimolecular fluorescence complementation of YFP in Xenopus oocytes. Scanning confocal microscopy images shows one-quarter of the oocyte either fluorescing (right image) or under bright field (left image). No background autofluorescence was observed in cells expressing untagged ILK1 (n = 6). C, The kinase domain of ILKs lacks several critical residues present in eukaryotic protein kinases. Alignment of the highly conserved amino acids within the kinase subdomains of the plant ILK family revealed higher conservation of kinase consensus residues in plants (black letters) compared to a representative metazoan ILK from Drosophila, which contains several substitutions (red letters). Asterisks indicate the residues mutated in this study. D, Autophosphorylation (ILK-P) and substrate phosphorylation (tubulin-P) of purified ILK1-cMyc with increasing concentrations of Mn2+ or Mg2+ cofactor visualized by autoradiography. Equal loading was determined by Ponceau staining. E, Autophosphorylation (ILK-P) and CML9 phosphorylation (CML9-P) with 5 mm Mn and 1 mm Ca, in the presence of increasing amounts of CML9-cMyc. Protein loading is visualized by western blot with an anticMyc antibody.