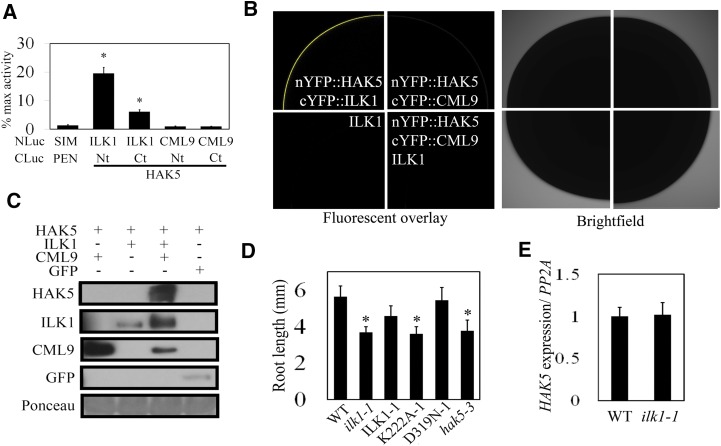
Figure 9.
ILK1 interacts with the HAK5 K+ uptake transporter and promotes growth in low K+ conditions. A, ILK1 and HAK5 cytosolic domains interact in Arabidopsis protoplasts. Luciferase signal resulting from interaction between bait (NLuc) and prey (CLuc) fusion proteins is normalized to the signal produced by a positive control pair (NLuc-H2A, CLuc-H2B). Four replicates from three independent experiments for each pair were used where * indicates significant difference from the negative control (NLuc-SIM, CLuc-PAN, P < 0.05). B, Images showing interaction of ILK1 with full-length HAK5 by bimolecular fluorescence complementation in Xenopus oocytes. The image shows the fluorescence (left panel) and bright field (right panel) confocal microscopy images of one-quarter of oocyte cells expressing the constructs indicated in each set. No interaction between nYFP::HAK5 and cYFP::CML9 was observed even upon coinjection with the untagged ILK1. Untagged ILK1 was transformed into cells as a negative control for background autofluorescence (n = 6). C, HAK5 accumulates when co-expressed with ILK1 and CML9. HAK5-GFP protein accumulation in the membrane fraction of N. benthamiana leaves was evaluated by western blot. HAK5 was co-expressed with GFP, CML-cMyc, or ILK1-cMyc, which are visible in the soluble fraction. Equal loading of total protein extract was verified by Ponceau stain. D, Primary root length under limiting K+ conditions (10 μm) 6 d after germination where * indicates significant difference from the wild type, P < 0.05. E, HAK5 expression is unaffected in ilk1-1 plants. The mean HAK5 expression from six replicates is presented, where expression is normalized to PP2A.