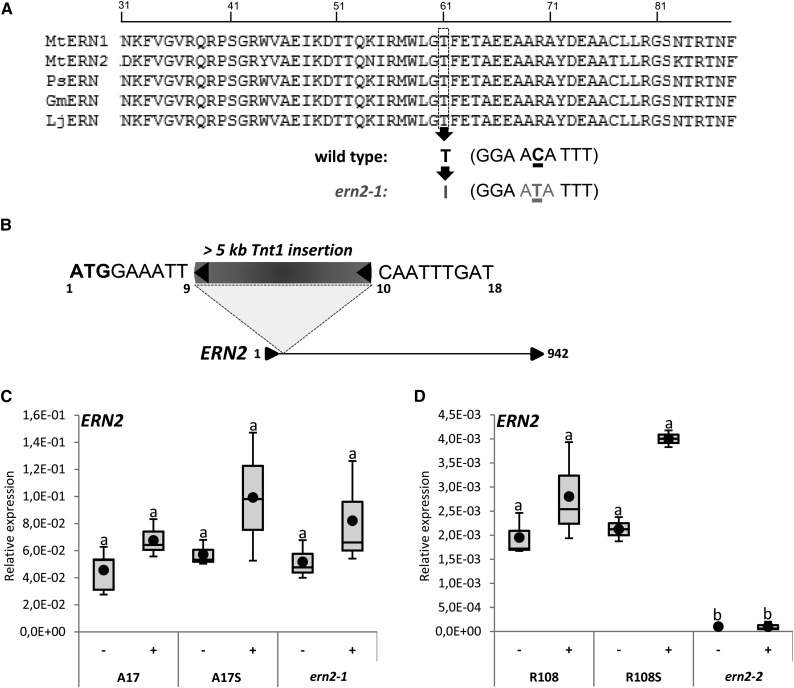
Figure 1.
Identification of ERN2 mutant alleles. A, Alignment of M. truncatula ERN1 (accession: EU038802) and ERN2 (accession: EU038803) DNA binding domains with closest Pisum sativum (PsERN, EF396329), Glycine max (GmERN, XM_006604176), and L. japonicus (LjERN, AP006677.1) ortholog sequences using Clustal W (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The conserved Thr (residue T61 in MtERN2) is replaced by an Ile (I) in the ern2-1 EMS mutant. B, The ern2-2 line is a knockout mutant with an approximately 5.3-kb Tnt1 retrotransposon inserted between nucleotides 9 and 10 downstream of the ERN2 start codon (position 1). Nucleotide sequences flanking the Tnt-1 insertion are indicated. The stop codon is at position 942. C and D, QRT-PCR analyses of ERN2 transcript levels in total RNA samples from M. truncatula roots treated for 6 h with water (−) or 10−9 m NF solution (+). RNA was extracted from wild type (A17 and R108), wild-type sibling (A17S and R108S), and ern2-1 (A17 background) and ern2-2 (R108 background). Box plots represent relative expression levels of two to four independent biological experiments after normalization against reference transcript levels. Box plots represent first and third quartile (horizontal box sides), minimum and maximum (outside whiskers). Black circles depict mean values. One-way ANOVA followed by a Tukey honest significant difference (HSD) test of the values was performed (in C, P > 0.05; in D, P < 0.001). Classes sharing the same letter are not significantly different.