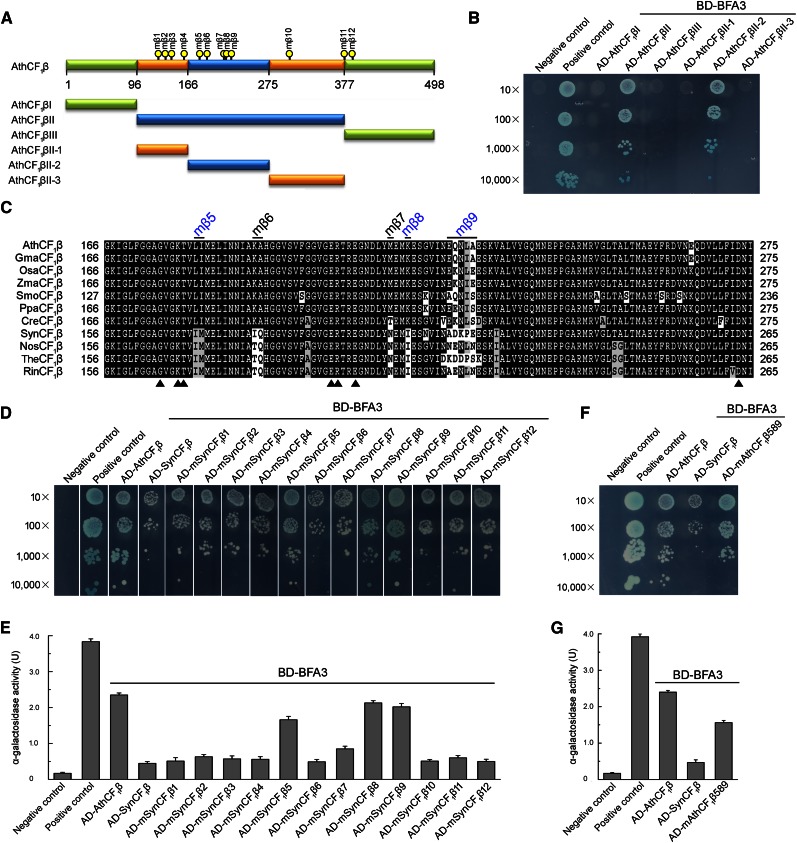
Figure 9.
Mapping the critical residues of CF1β required for the interaction with BFA3. A, Diagram of CF1β and various deletions (I, II, III, II-1, II-2, and II-3) of CF1β. The positions of the point mutation (mβ1-mβ12) are indicated by yellow dots. B, Yeast two-hybrid analysis of the interaction between BFA3 and various deletions of AthCF1β. C, Amino acid sequence alignment of the interacting region (AthCF1βII-2) corresponding to amino acid residues 166 to 275 in Arabidopsis CF1β. The positions of the point mutation (mβ5-mβ9) are indicated. The residues for nucleotide binding in CF1β are indicated by closed triangles. Ath, Arabidopsis thaliana; Gma, Glycine max; Osa, Oryza sativa; Zma, Zea mays; Smo, Selaginella moellendorffii; Ppa, Physcomitrella patens; Cre, Chlamydomonas reinhardtii; Syn, Synechocystis sp. PCC 6803; Nos, Nostoc sp. PCC 7120; The, Thermosynechococcus elongatus BP-1; Rin, Richelia intracellularis. D, Yeast two-hybrid analysis of the interaction between BFA3 and SynCF1β variants. In these variants, the residues in SynCF1β were point mutated to the corresponding residues in AthCF1β and designated as mSynCF1β1- mSynCF1β12. E, Quantitative α-galactosidase assay. The strength of interaction was quantified by assaying α-galactosidase activity in yeast colonies using p-nitrophenyl α-d-galactopyranoside as substrate. Mean and SD of α-Gal activity were obtained from three independent colonies. F, Yeast two-hybrid analysis of the interaction between BFA3 and the AthCF1β589 variant. In the AthCF1β589 variant, the residues in AthCF1β at the positions of mβ5, mβ8, and mβ9 (A and C) were point mutated to the corresponding residues in SynCF1β. G, Quantitative α-galactosidase assay performed as in E.