Gain of an autoregulatory site in one paralog led to the divergence of two duplicate genes in the time, space, and level of expression and regulation of one paralog by the other.
Abstract
How genes change their expression patterns over time is still poorly understood. Here, by conducting expression, functional, bioinformatic, and evolutionary analyses, we demonstrate that the differences between the Arabidopsis (Arabidopsis thaliana) APETALA1 (AP1) and CAULIFLOWER (CAL) duplicate genes in the time, space, and level of expression were determined by the presence or absence of functionally important transcription factor-binding sites (TFBSs) in regulatory regions. In particular, a CArG box, which is the autoregulatory site of AP1 that can also be bound by the CAL protein, is a key determinant of the expression differences. Because of the CArG box, AP1 is both autoregulated and cross-regulated (by AP1 and CAL, respectively), and its relatively high-level expression is maintained till to the late stages of sepal and petal development. The observation that the CArG box was gained recently further suggests that the autoregulation and cross-regulation of AP1, as well as its function in sepal and petal development, are derived features. By comparing the evolutionary histories of this and other TFBSs, we further indicate that the divergence of AP1 and CAL in regulatory regions has been markedly asymmetric and can be divided into several stages. Specifically, shortly after duplication, when AP1 happened to be the paralog that maintained the function of the ancestral gene, CAL experienced certain degrees of degenerate evolution, in which several functionally important TFBSs were lost. Later, when functional divergence allowed the survival of both paralogs, CAL remained largely unchanged in expression, whereas the functions of AP1 were gradually reinforced by gains of the CArG box and other TFBSs.
The expression pattern is one of the most important attributes of a gene. Understanding how the expression pattern of a gene is precisely determined and changes over time is key to understanding the nature of organismal development and evolution. In the past few decades, based on studies of model organisms, much has been learned about the molecular basis of gene regulation (Davidson, 2006; Arthur, 2011). Yet, it remains largely unclear how, why, to what extent, and under which conditions genes change their expression patterns. One reason for this is that expression itself is a complex process, or state, that requires measurements and descriptions from different angles, such as time, space, amount, and type (Arthur, 2011). Another reason is the difficulty of conducting appropriate experiments and analyses to determine the exact contribution of each evolutionary change to the differences in expression pattern (Wittkopp and Kalay, 2012; Hardison and Taylor, 2012). Nevertheless, based on theoretical and empirical investigations, several principles have emerged regarding the molecular basis of expression divergence (Prud’homme et al., 2007; Gordon and Ruvinsky, 2012; Romero et al., 2012). For example, it has been suggested that evolutionary changes in the expression pattern of a gene may be caused by alterations in cis-regulatory elements (CREs) or transcription environment, or both, although the relative contributions of the two mechanisms are usually difficult to determine (Wray et al., 2003). It has also been reported that while transcription environment itself is evolving all the time, changes of CREs have played important roles in shaping the expression patterns of genes (Wittkopp et al., 2004; Wray, 2007; Wittkopp and Kalay, 2012). Many studies also tried to determine the tempo, mode, and mechanisms of CRE evolution (Wittkopp and Kalay, 2012; Gordon and Ruvinsky, 2012; Villar et al., 2014), yet the available data are still insufficient for a general picture.
Interestingly, compared with orthologs from different species, paralogs from the same species are better systems for studying the tempo, mode, and mechanisms of CRE evolution, for three reasons. First, paralogs have evolved under the same transcription environment, so that most, if not all, of the differences in expression pattern may be attributed to changes in CREs (Li et al., 2005). Second, paralogs from the same model species can be compared, analyzed, or even manipulated with ease, thereby avoiding the difficulties of conducting interspecies comparisons (Kleinjan et al., 2008; Schauer et al., 2009). Third, and most importantly, the evolutionary fates of duplicate genes have been investigated extensively in the past few decades, based on which a few models have been proposed (Ohno, 1970; Force et al., 1999; He and Zhang, 2005; Moore and Purugganan, 2005; Innan and Kondrashov, 2010). In general, these models are both elegant and powerful, being able to explain the evolutionary fates of almost all duplicate genes. The problem, however, is that they mainly consider the consequences rather than the process of duplicate gene evolution (Innan and Kondrashov, 2010). In addition, none of these models take into consideration the cross-regulation between duplicate genes, and paralogs were generally assumed to evolve more or less independently (Innan and Kondrashov, 2010; Baker et al., 2013; Dhar et al., 2014; Rogozin, 2014). In reality, many duplicate genes are parts of the same regulatory network, in which the expression and function of one copy are dependent on those of the other, or vice versa (Kafri et al., 2006; Sémon and Wolfe, 2007; Conant et al., 2014). Therefore, a careful study of the molecular basis of duplicate gene evolution in expression pattern will not only uncover the tempo, mode, and mechanisms of CRE evolution but also shed new light on the evolution of the corresponding regulatory network.
Arabidopsis (Arabidopsis thaliana) APETALA1 (AP1) and CAULIFLOWER (CAL) are a pair of duplicate genes generated through a whole-genome duplication event within the flowering plant family Brassicaceae (Lawton-Rauh et al., 1999; Shan et al., 2007; Wang et al., 2012). As members of the MADS box gene family, both AP1 and CAL code for MIKC-type MADS domain-containing transcription factors and participate in plant development (Mandel et al., 1992; Bowman et al., 1993; Kempin et al., 1995; Ferrándiz et al., 2000; Han et al., 2014). Like many other duplicate gene pairs of the MADS box gene family, AP1 and CAL have diverged considerably in expression pattern (Supplemental Fig. S1). Specifically, AP1 is expressed mainly in floral primordia and developing sepals and petals, and the expression levels are generally high. Inactivation of AP1 caused the conversion of sepals into bracts and the concomitant formation of additional flowers in the axes of the bracts (Irish and Sussex, 1990; Mandel et al., 1992; Bowman et al., 1993), suggesting that AP1 not only determines the identity of the floral meristem but also specifies the identities of sepals and petals (Coen and Meyerowitz, 1991; Theissen, 2001). Unlike AP1, whose expression cannot be detected until floral meristems are formed, CAL expression can be detected even in seedlings (William et al., 2004), suggestive of early functioning. CAL is also expressed in floral meristems and developing sepals and petals, but the expression levels are relatively low (Kempin et al., 1995). Inactivation of CAL alone did not cause any obvious phenotypic change, while silencing of CAL in the ap1 background enhanced the phenotype of the plant, suggesting that CAL may have redundant function with AP1 (Bowman et al., 1993). The observation that the expression of AP1 decreased significantly in the young inflorescences of the ap1 cal double mutant but not in stage 1 and 2 flowers of the ap1 single mutant further led to the proposal that AP1 is positively regulated by CAL at the very early stage of flower development (Bowman et al., 1993). Clearly, as a pair of duplicate genes, AP1 and CAL have diverged considerably in the time, space, and level of expression and can be an excellent system for the study of the molecular basis of expression evolution.
Considerable progress has been made in understanding the mechanisms underlying the differences between AP1 and CAL. Up to now, it has been shown that (1) the protein products of the two genes, AP1 and CAL, have redundant but slightly differentiated functions, being able to interact with different numbers and sets of partners (Riechmann et al., 1996a, 1996b; Pelaz et al., 2001; de Folter et al., 2005; Kaufmann et al., 2005; Smaczniak et al., 2012); (2) the amino acid differences in the K and C regions of the AP1 and CAL proteins are responsible for their differences in function, whereas the M and I regions, which play key roles in binding to CREs of downstream genes, are functionally indistinguishable (Alvarez-Buylla et al., 2006); and (3) the expression of the AP1 and CAL genes is precisely controlled by many regulators, of which the vast majority are transcription factors (i.e. trans-regulatory elements; Wagner et al., 1999; Wigge et al., 2005; Saddic et al., 2006; Sundström et al., 2006; Kaufmann et al., 2009; Mathieu et al., 2009; Wang et al., 2009; Yamaguchi et al., 2009; Xu et al., 2010; Yant et al., 2010; Pastore et al., 2011). Several transcription factor-binding sites (TFBSs) have also been identified in the regulatory regions of AP1 and CAL, and functional studies indicate that their relative contributions to expression vary considerably (William et al., 2004; Wigge et al., 2005; Saddic et al., 2006; Sundström et al., 2006; Kaufmann et al., 2009, 2010; Mathieu et al., 2009; Wang et al., 2009; Yamaguchi et al., 2009; Yant et al., 2010; Benlloch et al., 2011; Pastore et al., 2011; Wuest et al., 2012). Despite this rapid progress, it remains unclear how AP1 and CAL have diverged in the time, space, and level of expression, how AP1 has acquired its function in sepal and petal identities, and how CAL has become a regulator of AP1.
In this study, by conducting a series of expression, functional, bioinformatic, and evolutionary analyses, we determine the molecular basis and evolutionary dynamics of the expression divergence between AP1 and CAL. We demonstrate that the differences between AP1 and CAL in the time, space, and level of expression were caused by gains and losses of functionally important TFBSs. We also show that the gains and losses of TFBSs along the lineages leading to the two paralogs have been quite dynamic and asymmetric, which, in turn, suggests that the divergence of duplicate genes in expression pattern is usually a complex process that cannot be easily depicted by simple empirical models. Our results provide new insights into the tempo, mode, and mechanisms of CRE evolution and highlight the necessity of conducting systematic experiments to understand the underlying mechanisms of duplicate gene evolution.
RESULTS
Differences in the Time, Space, and Level of Expression
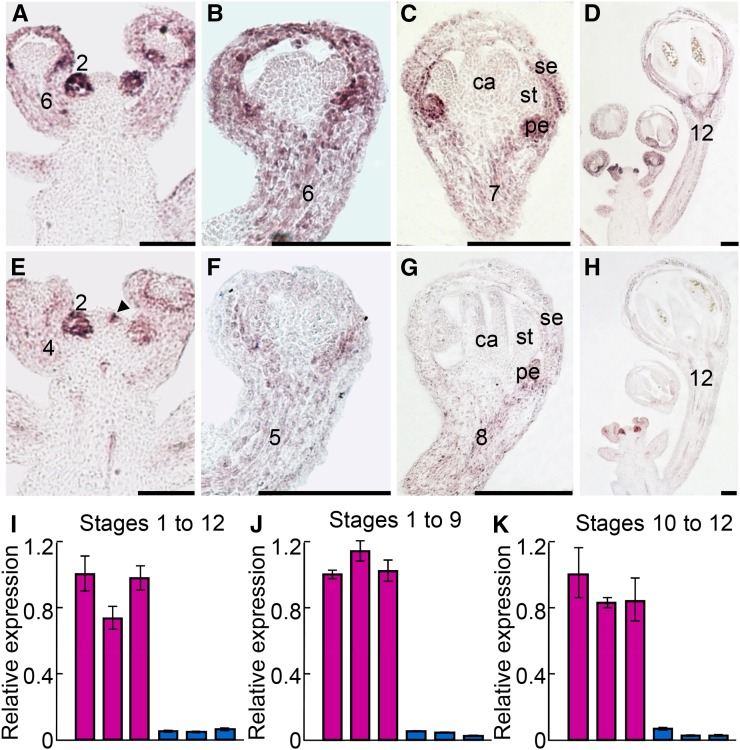
The expression patterns of AP1 and CAL were investigated in several studies (Mandel et al., 1992; Bowman et al., 1993; Gustafson-Brown et al., 1994; Kempin et al., 1995), yet the results are not completely consistent or comparable because different authors focused on different stages of flower development and because the resolution of the images was not always high. To get a clear portrait of the expression patterns of the two genes, we performed detailed in situ hybridization and quantitative real-time reverse transcription (qRT)-PCR analyses. We found that AP1 is strongly expressed in floral meristems (i.e. stage 2 flowers) and moderately expressed in developing sepals and petals in stage 3 to 12 flowers (Fig. 1, A–D). The expression pattern of CAL is very similar to that of AP1 but has three interesting differences (Fig. 1, E–K). First, its expression levels in floral meristems and developing sepals and petals are obviously lower than those of AP1, no matter which stage of flower development is considered and which method is used to measure. Second, roughly from stage 4 on, the expression of CAL decreases dramatically and vanishes eventually, while that of AP1 persists to late stages of flower development, with strong signals being detectable even in near-mature (stage 12) flowers. Third, CAL is also expressed in the cells underneath the floral buttress, whereas AP1 is not, suggesting that the expression of CAL is slightly earlier than that of AP1. Taken together, these results confirm that AP1 and CAL have diverged considerably in the time, space, and level of expression.
Figure 1.
Expression patterns of AP1 and CAL. A to H, Results of in situ hybridization for AP1 (A–D) and CAL (E–H). ca, Carpel; pe, petal; se, sepal; st, stamen. Stages of flower development were determined as described (Smyth et al., 1990). The arrowhead points to the cells underneath the floral buttress. Bars = 100 µm. I to K, Results of qRT-PCR for AP1 (purple) and CAL (blue) in inflorescences bearing flowers of stages 1 to 12 (I), 1 to 9 (J), and 10 to 12 (K). For each gene, three biological replicates were conducted, and error bars indicate the sd of three technical replicates of each biological replicate.
Differences in the Number and Type of TFBSs
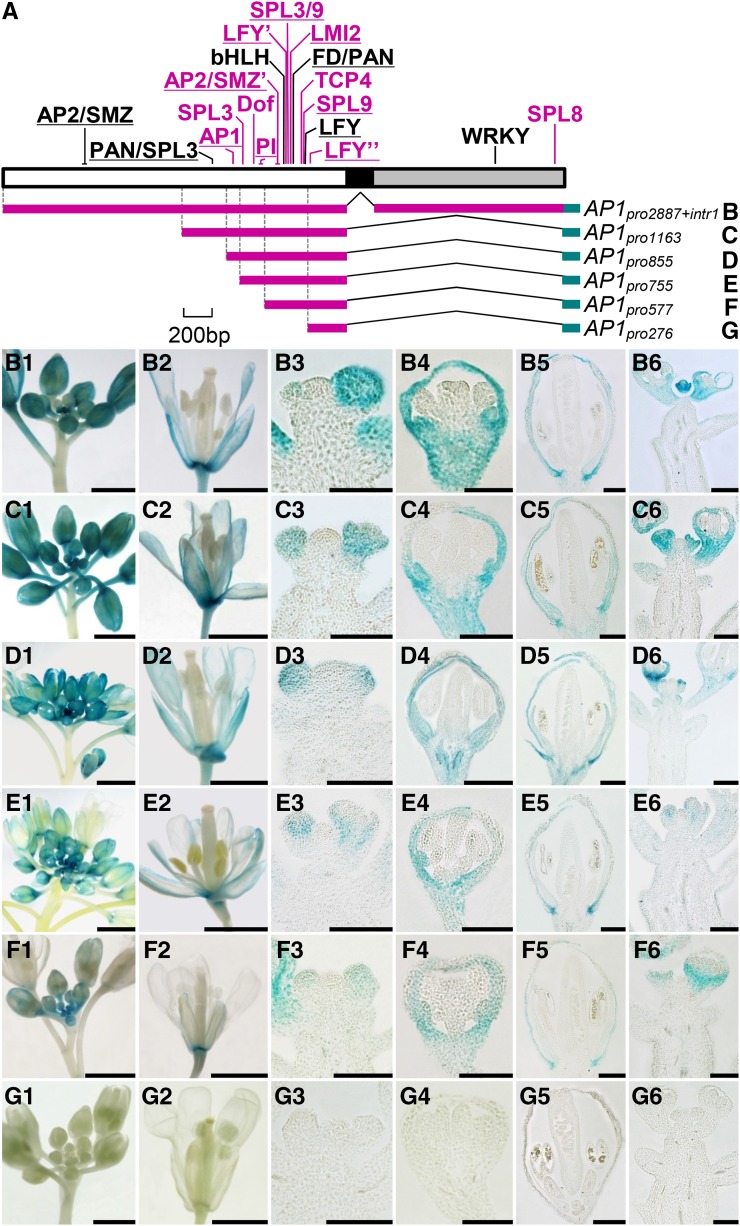
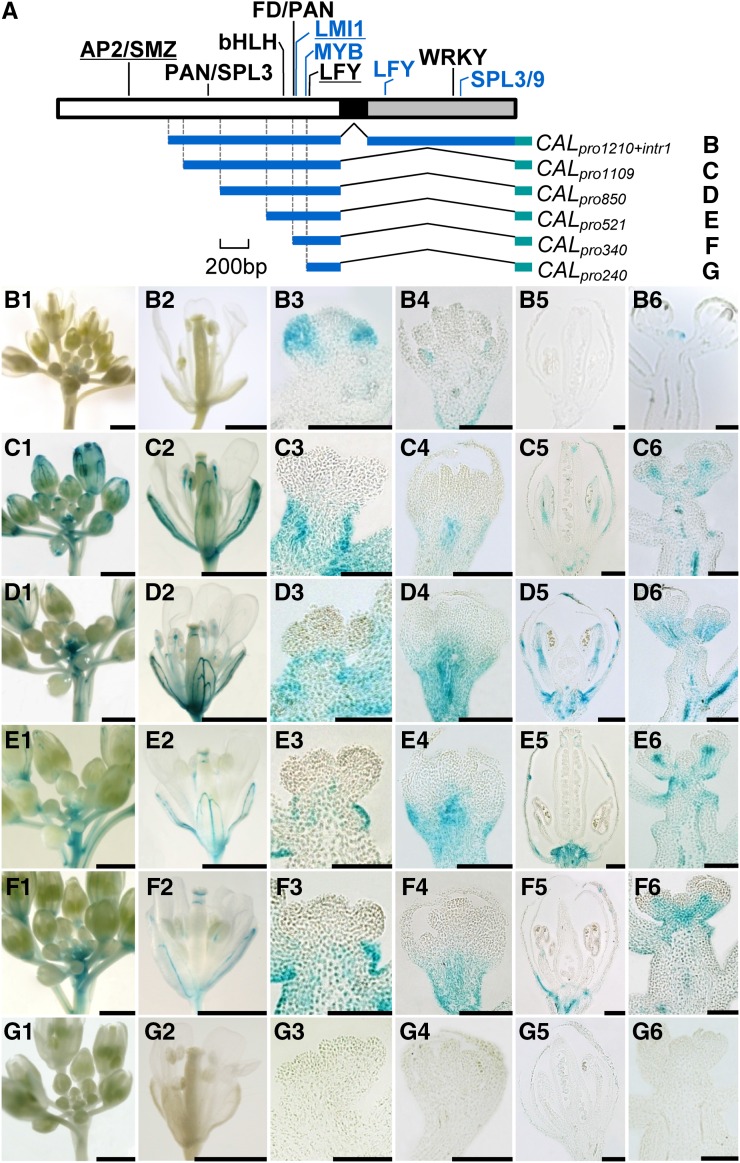
To understand the mechanisms by which AP1 and CAL have diverged in expression pattern, we first compared the genomic sequences of the two genes. For AP1, a 6,946-bp region was investigated, which covers all eight exons and seven introns plus 2,900 bp upstream of the translation start site and 519 bp downstream of the stop codon. Using the public resources for TFBS prediction and referring to published results (for details, see “Materials and Methods”), we identified 19 relatively reliable TFBSs: 16 in the promoter region and three in the first intron (Fig. 2A; Supplemental Figs. S2–S4). Twelve of these TFBSs have already been identified and functionally characterized in previous studies, suggestive of reliability (Supplemental Table S1); the remaining seven have not been confirmed by experiments, but the available data suggest that they are reliable because their sequences are identical, or nearly so, to those identified before and because corresponding trans-regulatory elements have been reported to function in relevant pathways (Supplemental Table S1). Using the same strategy, we identified 12 TFBSs in the 5,756-bp genomic region of CAL: seven in the promoter region and five in the first intron (Fig. 3A; Supplemental Figs. S2–S4). Of these TFBSs, only three have been functionally characterized, suggestive of the scarcity of studies on CAL relative to AP1 (Supplemental Table S1). Notably, only six TFBSs are shared by CAL and AP1 (Figs. 1 and 2; Supplemental Figs. S3 and S4), suggesting that the two genes have diverged considerably in CRE constitution. Since independent gains of exactly the same TFBS in different evolutionary lineages occur rarely (Wittkopp and Kalay, 2012), it is very likely that the shared TFBSs have existed in the most recent common ancestor of AP1 and CAL; then, after gene duplication, they were retained in both genes. For understanding the mechanisms underlying the divergence of AP1 and CAL in expression pattern, however, these shared TFBSs are not very useful.
Figure 2.
Regulatory regions of AP1 and their contributions to expression pattern. A, Predicted TFBSs in the promoter (white box), the first exon (black box), and the first intron (gray box). TFBSs in black are those shared by AP1 and CAL, whereas those in purple are AP1 specific. Experimentally confirmed TFBSs are underlined. B to G, Genomic regions used to drive the expression of GUS and the resultant expression patterns. Bars = 1 mm in columns 1, 2, and 6 and 100 µm in columns 3 to 5.
Figure 3.
Regulatory regions of CAL and their contributions to expression pattern. A, Predicted TFBSs in the promoter (white box), the first exon (black box), and the first intron (gray box). TFBSs in black are those shared by AP1 and CAL, while those in blue are CAL specific. Experimentally confirmed TFBSs are underlined. B to G, Genomic regions used to drive the expression of GUS and the resultant expression patterns. Bars = 1 mm in columns 1, 2, and 6 and 100 µm in columns 3 to 5.
Contributions of Different Regulatory Regions to Expression Pattern
To gain some insights into the roles of the identified TFBSs in gene expression, we performed a series of transgenic experiments. Transformable constructs containing different lengths of genomic regions (Fig. 2A) were first fused with the GUS gene (a reporter) and then introduced into Arabidopsis plants. When a construct (i.e. AP1pro2887+intr1) including the promoter and the first intron of AP1 was used, an expression pattern of GUS that completely matches that of AP1 was observed (Fig. 2B). This suggests that the GUS system worked well here and that the promoter and the first intron contain most, if not all, of the information needed for the normal expression of AP1. The fact that the same results were obtained when the AP1pro1163 and AP1pro855 constructs were used (Fig. 2, C and D), however, suggests that the contributions of the first half of the promoter (from −2,887 to −855) and the first intron are negligible. Interestingly, when a construct (i.e. AP1pro755) containing a shorter region was used, a slight but obvious decrease in expression level was observed (Fig. 2E), suggesting that the region spanning from −855 to −755 contains the CREs that enhance the expression of AP1. The observations that AP1pro577 gave the same results as AP1pro755 (Fig. 2F), whereas no GUS signal could be detected for AP1pro276 (Fig. 2G), suggest that the region spanning from −577 to −276 contains the basic information for the spatiotemporal expression of AP1.
We also applied the same strategy to CAL (Fig. 3A). In plants expressing the longest construct, CALpro1210+intr1, the expression pattern of GUS is completely congruent with that of CAL (Fig. 3B), suggesting that the region covering the promoter and the first intron contains almost all the CREs needed for the normal expression of CAL. In plants expressing CALpro1109, no GUS signal could be detected in the floral meristem, whereas the signals in developing sepals and petals become stronger (Fig. 3C). This suggests that the region spanning from −1,210 to −1,109 or the first intron, or both, are critical for the repression of CAL expression in pedicel/stem and the promotion of CAL expression in floral meristem. The observations that GUS signals in plants expressing CALpro850, CALpro521, or CALpro340 are not very different from that of CALpro1109 (Fig. 3, D–F), however, imply that the region spanning from −1,109 to −340 is not very important for CAL expression, in spite of the existence of several TFBSs. Alternatively, this region may have been involved in CAL expression by coordinating with the TFBSs located within the region spanning from −1,210 to −1,109 and/or the first intron. As in AP1, the promoter region spanning from −340 to −240 may provide the basic information for the spatiotemporal expression of CAL, because no GUS signal could be detected in the CALpro240 plants (Fig. 3G).
Functions of an Autoregulatory Site
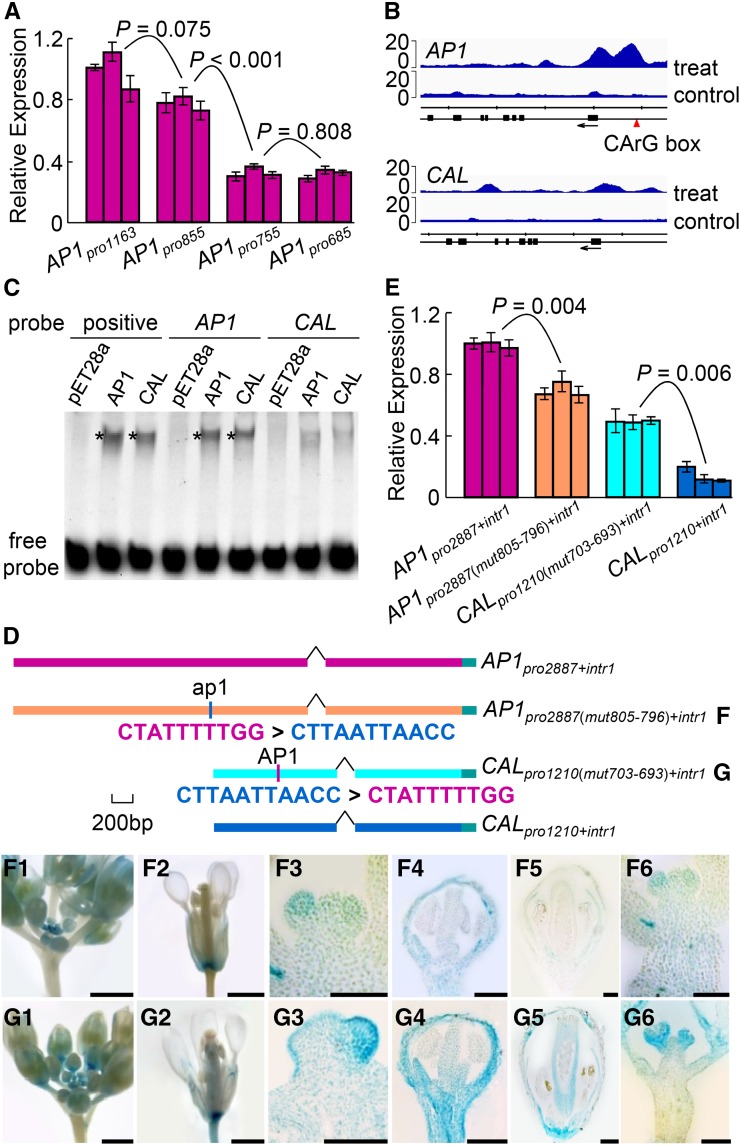
It is interesting that the region spanning from −855 to −755 is critical for the expression level of AP1 (Fig. 2, D and E). To understand the function of this region, we measured the expression levels of the GUS gene in plants expressing four different constructs (i.e. AP1pro1163, AP1pro855, AP1pro755, and AP1pro685). We found that the expression level was reduced to about half when the region spanning from −855 to −755 was excluded (Fig. 4A), suggesting that this region is indeed important in expression level maintenance. To figure out the mechanism behind this, we inspected this region carefully and encountered a CArG box, which, according to recent chromatin immunoprecipitation sequencing (ChIP-seq) studies of MADS box proteins (Kaufmann et al., 2009, 2010; Zheng et al., 2009; Deng et al., 2011; Immink et al., 2012; Wuest et al., 2012; Gregis et al., 2013; Ó’Maoiléidigh et al., 2013; Posé et al., 2013), is a putative binding site of the AP1 protein and its interacting partner, SEPALLATA3. Therefore, we hypothesized that this CArG box may have led to the formation of an autoregulatory loop through which the expression of AP1 is maintained at high levels to later stages of sepal and petal development; CAL does not have this CArG box and, thus, is expressed at very low levels in near-mature sepals and petals.
Figure 4.
Function of the AP1-binding CArG box. A, qRT-PCR results showing GUS signals in plants expressing four different constructs. For each construct, three independent transgenic lines were conducted. Error bars indicate the sd of three technical replicates. B, ChIP-seq results, reanalyzed from the data of Kaufmann et al. (2010), showing the AP1-binding regions around the AP1 (top) and CAL (bottom) genes. Sequenced reads from two biological replicates were combined and plotted as normalized read coverage on the vertical axis against the genomic location along the horizontal axis. Treat and control represent 35S:AP1-GR ap1-1 cal-1 plants treated and untreated, respectively, with dexamethasone-containing solution. Arrows indicate gene orientations. The scale division corresponds to 1,000 nucleotides. C, EMSA showing that the binding affinities of the AP1 and CAL proteins to the CArG box-containing AP1 probe are much stronger than to the CArG box-lacking CAL probe. Positive probe contains a canonical AP1-binding CArG box that has been verified in vitro (Riechmann et al., 1996b). pET28a represents a negative control in which the in vitro translation assay was programmed with the empty pET28a vector. Asterisks indicate the positions of the protein-DNA complexes. D, GUS constructs used to determine the functions of the CArG box. AP1pro2887(mut805-796)+intr1 and CALpro1210(mut703-693)+intr1 are two constructs in which the CArG box of AP1 was swapped with a piece of non-CArG box sequence of CAL in the corresponding position. E, qRT-PCR results showing GUS expression in plants expressing the four constructs in D. F and G, GUS signals in plants expressing the AP1pro2887(mut805-796)+intr1 (F) and CALpro1210(mut703-693)+intr1 (G) constructs. Bars = 1 mm in columns 1, 2, and 6 and 100 µm in columns 3 to 5.
To test this hypothesis, we first reanalyzed the published ChIP-seq data (Kaufmann et al., 2010) and observed two obvious AP1-binding peaks in the promoter region of AP1; the corresponding region of CAL, however, does not show such binding signals (Fig. 4B). We then performed electrophoretic mobility shift assay (EMSA) analyses and found that the binding affinities of the AP1 and CAL proteins to the CArG box-containing AP1 probe are clearly stronger than those to the CArG box-lacking CAL probe (Fig. 4C). We also made two constructs in which the CArG box of AP1 was swapped with a stretch of non-CArG box sequence in the corresponding region of CAL (Fig. 4D). As expected, the expression level of GUS in AP1pro2887(mut805-796)+intr1 is significantly lower than that in AP1pro2887+intr1 (two-tailed Student’s t test, P = 0.004; Fig. 4E), while that in CALpro1210(mut703-693)+intr1 is significantly higher than that in CALpro1210+intr1 (two-tailed Student’s t test, P = 0.006; Fig. 4E). Notably, when the CArG box was removed from AP1pro2887+intr1, the expression pattern became more similar to that of CAL than to AP1: signals were initially detected in floral primordia and developing sepals and petals but eventually vanished in near-mature flowers (Fig. 4F). Conversely, when the CArG box was added to CALpro1210+intr1, the expression pattern became more similar to that of AP1 than to CAL: signals were detectable throughout flower development, even in near-mature sepals and petals (Fig. 4G). Taken together, these results suggest that the CArG box is an autoregulatory site of AP1 that can also be bound by CAL, thereby functioning to maintain the relatively high levels of AP1 expression in near-mature sepals and petals.
Origin of the Autoregulatory Site
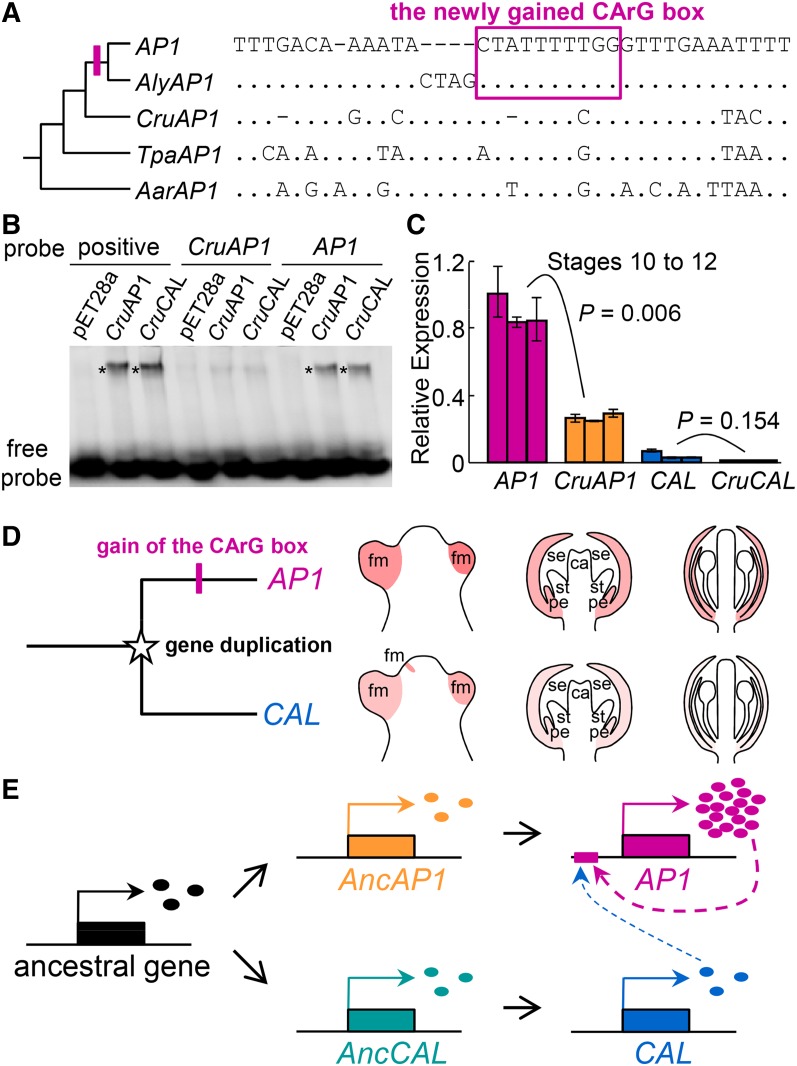
To understand the evolutionary history of the CArG box, we obtained orthologs of AP1 and CAL in Arabidopsis lyrata, Capsella rubella, Thellungiella parvula, and Aethionema arabicum (i.e. AlyAP1, AlyCAL, CruAP1, CruCAL, TpaAP1, TpaCAL, AarAP1, and AarCAL, respectively), as well as the AP1/CAL-like genes in Tarenaya hassleriana and Carica papaya, and compared their regulatory regions (Supplemental Fig. S5; Supplemental Table S2). We found that the aforementioned CArG box is also present in AlyAP1 and that the sequence is completely identical to that of AP1 (Fig. 5A); in other genes, however, no such CArG box is recognizable, although similar sequences with very few mismatches do exist in the corresponding regions (Fig. 5A). In CruAP1, for example, there is a similar sequence that has an alignment gap in the third position and a C rather than a T in the eighth position. Similarly, in TpaAP1 and AarAP1, there are similar sequences that possess two nucleotide differences in this otherwise highly conserved region (Fig. 5A). Previous studies have shown that, when nucleotides at these positions were removed or mutated, the resulting sequences would no longer be functional, unable to interact with relevant MADS box proteins (Huang et al., 1996). Therefore, it is very likely that the corresponding sequences in CruAP1, TpaAP1, and AarAP1 are not CArG boxes.
Figure 5.
Origin of the AP1-binding CArG box and its consequences. A, Alignment of the corresponding regions in a phylogenetic framework, which suggests that the CArG box was gained in the ancestor of AP1 and AlyAP1, likely through modification of a preexisting non-CArG box sequence. Dots represent the same nucleotides as those in AP1, whereas dashes indicate alignment gaps. B, EMSA showing the binding ability of CruAP1 and CruCAL in vitro. Positive probe contains a canonical AP1-binding CArG box that has been verified in vitro (Riechmann et al., 1996b). pET28a represents a negative control in which an in vitro translation assay was programmed with the empty pET28a vector. Asterisks show the positions of the DNA-protein complexes. C, qRT-PCR results showing the relative expression levels of AP1 and CruAP1 in inflorescences bearing flowers of stages 10 to 12. For each gene, three biological replicates were conducted, and error bars indicate the sd of three technical replicates. D, Cartoon showing the divergence of AP1 and CAL in expression pattern. The gene duplication giving rise to AP1 and CAL occurred before the origin of the Brassicaceae, while the gain of the CArG box happened after the divergence of Arabidopsis from Capsella. Because of the gain of the CArG box, AP1 and CAL diverged in the time, space, and level of expression. ca, Carpel; fm, floral primordia; pe, petal; se, sepal; st, stamen. E, Model depicting the gain of the CArG box and the formation of a regulatory cascade involving AP1 and CAL. AncAP1, Ancestor of the AP1 orthologs; AncCAL, ancestor of the CAL orthologs; ovals represent proteins of the corresponding genes.
To test this hypothesis, we first conducted EMSA experiments. We found that proteins of both CruAP1 and CruCAL can strongly bind to the CArG box-containing AP1 probe, whereas their binding to the CArG box-like sequence of CruAP1 is rather weak (Fig. 5B). This confirms that the CArG box-like sequences of CruAP1 are unlikely to be functional. We then compared the expression level of CruAP1 with that of AP1. Theoretically, because CruAP1 does not have the CArG box, it would not be autoregulated and thus its expression level should be lower than that of AP1. Indeed, when flowers at the same developmental stage were compared, the expression level of CruAP1 was markedly lower than that of AP1 (Fig. 5C); the expression levels of CruCAL and CAL, however, did not show significant differences (Fig. 5C). This suggests that the CArG box-like sequence of CruAP1 is not functionally equivalent to the real CArG box. Taken together, these results not only confirm the importance of the CArG box in AP1 expression but also indicate that the particular CArG box has been gained through modification of a preexisting non-CArG box sequence in the ancestor of AP1 and AlyAP1 and, as a result, contributed to the differences between AP1 and CAL in the time, space, and level of expression (Fig. 5, D and E).
Gains and Losses of Other TFBSs
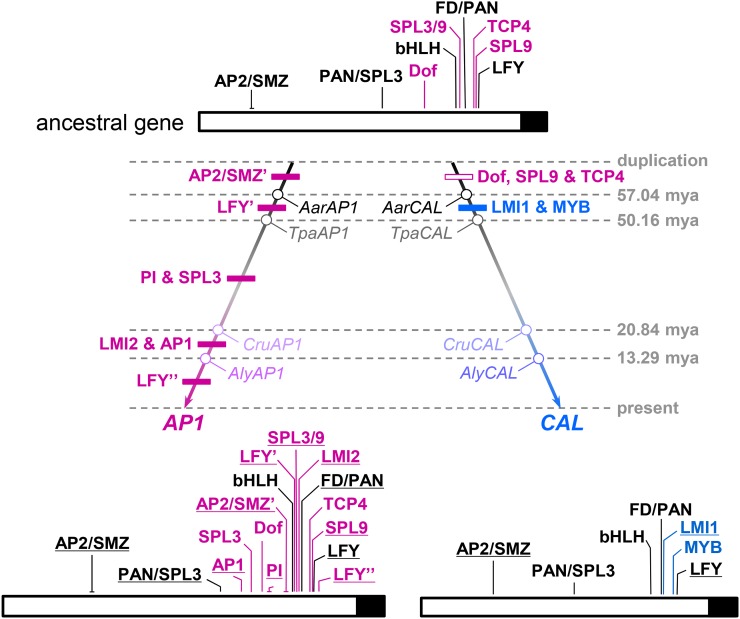
To understand the evolutionary context of the CArG box origination, we also tried to trace the evolutionary histories of other TFBSs. However, because exact functions of the predicted TFBSs in other species remain to be determined, we only considered the evolutionary changes of the sequences per se, with special attention being paid to functionally important nucleotide sites. Meanwhile, because the evolutionary histories of the TFBSs within the first intron turned out to be extremely difficult to elucidate, we focused instead on those located in the promoter region. We found that at least nine TFBSs existed in the most recent common ancestor of AP1 and CAL (Fig. 6; Supplemental Fig. S5; Supplemental Table S2), among which five have been retained by both genes. After gene duplication, seven and two new TFBSs were gained along the lineage leading to AP1 and CAL, respectively. Three TFBSs were also lost in the lineage leading to CAL, whereas no TFBS-loss events could be deduced for the lineage leading to AP1. Interestingly, while gains of TFBSs along the AP1 lineage occurred more or less gradually, losses of three TFBSs along the CAL lineage all occurred at the very early stages of postduplication evolution, immediately followed by the gains of two new TFBSs (Fig. 6). This suggests that AP1 and CAL not only gained/lost different sets of TFBSs but also experienced different modes of evolution.
Figure 6.
TFBS evolution along the lineages leading to AP1 and CAL. In the ancestral gene, TFBSs shared by AP1 and CAL are in black, whereas those specific to AP1 are in purple. Experimentally confirmed TFBSs are underlined. TFBSs gained and lost during different evolutionary stages are indicated in the corresponding positions of the phylogenetic tree by black and white boxes, respectively. The loss of the binding site of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE3/9 (SPL3/9) along the CAL lineage is not shown, however, because its exact position is still uncertain. The ages (mya, million years ago) of nodes indicated by gray numbers are based on Beilstein et al. (2010). For details, see Supplemental Figure S5.
Notably, most of the TFBSs gained or lost along the lineages leading to AP1 and CAL resulted from modifications of local sequences rather than from translocations of preexisting ones, because similar sequences can still be found in the corresponding regions of the orthologous and/or paralogous genes (Supplemental Fig. S6). The LEAFY (LFY)-binding site located between −255 and −250 bp of the AP1 promoter, for example, likely resulted from the substitution of an A with a G at the sixth position after the divergence between AP1 and AlyAP1. This, together with the observation that the TFBSs gained or lost at nearly the same evolutionary stage are usually located in different parts of the promoters (Fig. 6), further suggest that gains and losses of TFBSs have occurred separately rather than collectively or massively.
DISCUSSION
A Versatile CArG Box
In this study, by conducting extensive expression analyses, we first confirmed that AP1 and CAL have diverged in the time, space, and level of expression. Then, by comparing and functionally dissecting the regulatory regions of the two genes, we identified the portions that are responsible for the differences in expression pattern. We found that most of the differences in expression pattern can be explained by the presence or absence of certain portions of the regulatory regions, each of which contains functionally important TFBSs. In particular, a CArG box in the promoter region of AP1 seems to be a key to understanding the mechanisms that underlie the expression differences between AP1 and CAL. Replacement of the CArG box with a non-CArG box sequence in the AP1-based GUS construct led to decreased GUS expression in developing sepals and petals, whereas substitution of this non-CArG box sequence with the CArG box in the CAL-based GUS construct led to increased GUS expression in those organs. This, together with the fact that the CArG box can interact with proteins coded by AP1 and CAL, suggests that it is the autoregulatory site of AP1 that can also be bound by CAL. CAL and AP1, therefore, form a regulatory cascade in which AP1 is both autoregulated by itself and cross-regulated by CAL; it is for this reason that the relatively high levels of AP1 expression can be maintained till to the late stages of sepal and petal development. CAL does not have this CArG box (or any other TFBSs of this kind) and thus cannot be regulated directly by either AP1 or CAL, so that its expression is transient and low leveled. In addition, because of the formation of a regulatory cascade, as well as the slightly earlier expression of CAL than AP1, CAL promotes the expression of AP1 at the very early stage of flower development (Bowman et al., 1993; Gustafson-Brown et al., 1994; William et al., 2004). Up to now, although this last point still needs to be proved in vivo, it is already clear that, as a particular CRE, the CArG box is versatile, being able to cause considerable differences in the time, space, and level of expression.
It is interesting that this particular CArG box was gained recently, very likely before the divergence of Arabidopsis and A. lyrata but after the Arabidopsis-Capsella split (Fig. 5). This implies that the regulation of AP1 by AP1 itself and CAL, as well as the relatively high levels of expression in developing sepals and petals, are derived features shared by AP1 and AlyAP1 but not by CruAP1, TpaAP1, AarAP1, or the CAL orthologs. This is very interesting, because, unlike its counterparts in many other species, which are generally not involved in floral organ identity determination (Huijser et al., 1992; Taylor et al., 2002; Vrebalov et al., 2002; Litt, 2007), AP1 not only regulates the formation of floral primordia but also is involved in sepal and petal development. Presumably, it was the gain of this CArG box that enabled AP1 to extend its roles in sepal and petal development.
Autoregulation and Duplicate Gene Evolution
Our results also highlighted the importance of autoregulation in duplicate gene evolution. As a special type of regulation, autoregulation exists in all kinds of life forms and plays particularly important roles in maintaining the expression levels of genes (Crews and Pearson, 2009). Autoregulation can be positive or negative, direct or indirect, depending on the function of the genes (Crews and Pearson, 2009). In any case, duplication of a gene capable of autoregulation may lead to complex consequences, because the resultant duplicates would form a regulatory network (Studer et al., 1998; Czerny et al., 1999; Sémon and Wolfe, 2007; Lenser et al., 2009). Loss of the autoregulatory site, therefore, will cause the interruption of existing regulatory relationship(s) between genes. In the past, possibly because of its complexity, autoregulation has not been explored extensively in terms of its effects on duplicate gene evolution. Even in the limited published case studies, much attention has been paid to the effect of gene duplication on the maintenance of the regulatory relationships between genes (Teichmann and Babu, 2004; Sémon and Wolfe, 2007); the contribution of newly established autoregulation to duplicate gene evolution, however, remains largely unexplored.
In this study, it is clear that the gain of the CArG box not only enabled the divergence of AP1 and CAL in the time, space, and level of expression but also led to the formation of a regulatory cascade, thereby further splitting the function domains of the two genes. Meanwhile, because AP1 is both autoregulated and cross-regulated, its function is strengthened, with relatively high-level expression being maintained until the late stages of sepal and petal development. Without the CArG box, the expression level of AP1 would not be that high, and the high-level expression would not be maintained from floral meristem to developing and near-mature sepals and petals. Consistent with this, AP1 has evolved under more stringent functional constraint than CAL, as reflected by its relatively low dN/dS value (Lawton-Rauh et al., 1999). Apparently, gain of the autoregulatory site has enabled AP1 and CAL to arrive at a state that cannot be easily reached through many other mechanisms. Because independent origins of CREs through modifications of preexisting sequences are rather easy, it is possible that similar phenomena exist in other genes and other organisms, and more studies are needed to clarify this issue.
Contributions of Other TFBSs
In spite of its importance, the CArG box is unlikely to be the only CRE that determines the expression differences between AP1 and CAL. Direct evidence supporting this comes from the sequence-swapping experiments: when the CArG box of AP1pro2887+intr1 was replaced by a piece of non-CArG sequence, the expression of GUS decreased, but not down to the level in CALpro1210+intr1; when the non-CArG sequence of CALpro1210+intr1 was replaced by the CArG box, the expression of GUS increased, but not up to the level in AP1pro2887+intr1 (Fig. 4E). This suggests that the CArG box is only part of the story and that other TFBSs, especially those gained or lost along the lineages leading to the two genes, must have also been essential, although their exact contributions are still unclear. Indeed, of the recently gained TFBSs along the lineage leading to AP1, several have been shown to be functionally important. The LFY-binding site between positions −419 and −414, for example, has been shown to be critical for the initial expression of AP1, because deletion of it can cause a later response to photoperiodic induction (Benlloch et al., 2011). Similarly, the PISTILLATA (PI)-binding site, which is also a CArG box but spans from −603 to −595, is the CRE to which the AP3-PI heterodimer binds and represses AP1 expression (Sundström et al., 2006; Wuest et al., 2012). The fact that exclusion of a 100-bp-long promoter region and the first intron led to nearly complete loss of CAL expression in floral primordia further suggests that the differences between AP1 and CAL in the time, space, and level of expression were caused by multiple factors. Additional analyses are needed to elucidate the functions and contributions of the TFBSs in this region.
Interestingly, in both AP1 and CAL, a relatively short region seems to be sufficient for the basic expression: constructs lacking this region were generally not expressed anywhere. In CAL, this region spans from −340 to −240 and contains at least four TFBSs, whereas in AP1, the region spans from −450 to −310 and contains at least five TFBSs (Wigge et al., 2005; Kaufmann et al., 2009). Because the binding sites of FLOWERING LOCUS D (FD) and PERIANTHIA (PAN) are shared by the two genes, these results highlighted the importance of the FD and PAN proteins in AP1 and CAL expression. As members of the bZIP transcription factor family, both FD and PAN are key regulators of flower development, able to bind to the regulatory regions of AP1 and CAL and induce their expression (Wigge et al., 2005; Xu et al., 2010). FD can form a protein complex with FLOWERING LOCUS T, the florigen, to activate flower identity genes (Wigge et al., 2005), whereas PAN plays key roles in determining the number and position of floral organs, as inactivation of it led to the generation of pentamerous rather than the normally tetramerous flowers (Chuang et al., 1999). The fact that the binding sites of FD and PAN are largely overlapping and highly conserved further implies that their functions may be interdependent. Presumably, it is the FD/PAN-binding site, together with other TBFSs (e.g. LFY) in this region, that determines the on/off of the two genes, whereas the TFBSs in other regions adjust and fine-tune the time, space, and level of expression.
Dynamics of TFBS Evolution
It is interesting that the divergence of AP1 and CAL in expression has been markedly asymmetric. In the lineage leading to AP1, at least seven TFBSs were gained, while no loss event could be deduced. In the lineage leading to CAL, however, at least three and two TFBSs were lost and gained, respectively. Interestingly, while the gains of TFBSs along the AP1 lineage occurred more or less gradually during evolution, the losses and gains of TFBSs along the lineage leading to CAL all occurred at the early stages of postduplication evolution (Fig. 6); after that, CAL did not gain or lose any TFBS, while AP1 gained seven more TFBSs in a step-by-step manner. This suggests that, shortly after gene duplication, CAL experienced a period of degenerate evolution so that several functionally important TFBSs (i.e. Dof, TCP4, and SPL9) were lost. Consistent with this, CAL evolved under less stringent functional constraint than AP1 and even experienced an exonization event in the 5′ end of the third exon during roughly the same period (Supplemental Figs. S7 and S8). Presumably, it was the degenerate evolution of CAL that allowed the survival and additional diversification of both CAL and AP1.
Notably, of the TFBSs that were gained along the lineage leading to AP1, most have been shown to be functionally important, through which a regulatory network involving a handful of genes was formed (Parcy et al., 1998; Sundström et al., 2006; Mathieu et al., 2009; Yamaguchi et al., 2009; Kaufmann et al., 2010; Yant et al., 2010; Pastore et al., 2011). Within this network, some genes function as activators and others as repressors, so that the expression of AP1 is precisely regulated. Without these TFBSs, the regulation of AP1 would be as simple as that of CAL and the regulatory network specifying floral primordia would not be so sophisticated. In addition, because these TFBSs were gained gradually, it is very likely that, after gene duplication, when AP1 happened to be the one that maintained the function of the ancestral gene, its functions were reinforced further by gaining additional TFBSs. Thereafter, because of the continuous reinforcements, AP1 became one of the most important regulators of flower development. Clearly, the processes through which AP1 and CAL become diverged in expression pattern were both dynamic and asymmetric and can be regarded as an excellent model for duplicate gene evolution in regulatory regions.
Mechanisms of CRE Evolution
Several studies have attempted to summarize the mechanisms through which CREs may evolve, but no consensus has been reached (Wittkopp and Kalay, 2012; Villar et al., 2014). In this study, it is clear that the vast majority of the gained or lost TFBSs along the lineages leading to AP1 and CAL were the results of modifications of preexisting sequences (Fig. 5A; Supplemental Fig. S6). For the TFBSs whose evolutionary histories remain unclear, the possibility of modification also cannot be excluded. This suggests that modification (rather than translocation) of preexisting sequences may be a common means of CRE origination. Indeed, because the number of nucleotides is very large whereas CREs are generally short DNA pieces, it may not be very difficult for CREs to evolve through the modification of pre-existing sequences, if the time of evolution is sufficiently long and if the sequences with degenerate sites can also be recognized by corresponding transcription factors. The fact that the binding sites of the same transcription factor (such as LFY) can sometimes be found in the different, nonhomologous regions of paralogous genes (Fig. 6) further indicates the ease of TFBS origination and the fluidic nature of TFBS functioning. However, without detailed comparisons between closely related species within a phylogenetic framework, like what we have done in this study, it is very difficult to determine whether the TFBSs of the same sequence features and/or functional properties are homologous and how newly originated TFBSs were generated.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana; ecotype Columbia-0 [Col-0]) and Capsella rubella (accession 86IT1) were surface sterilized by treating with 70% (v/v) ethanol and 10% (v/v) hypochlorite and plated on one-half-strength Murashige and Skoog medium supplemented with 1% (w/v) Suc and 0.8% (w/v) agar. The plates were cold treated at 4°C for 3 to 4 d and then transferred to a standard growth room. After germination, seedlings were transferred to soil and grown under a 16-h-light (22°C)/8-h-dark (18°C) photoperiod and 60% relative humidity.
Identification and Evolutionary Analyses of TFBSs
Genomic sequences of the Arabidopsis AP1 and CAL genes were retrieved from The Arabidopsis Information Resource 10 (http://www.arabidopsis.org/). Putative TFBSs along the genomic sequences were first predicted with the help of AGRIS (http://arabidopsis.med.ohio-state.edu; Yilmaz et al., 2011) and AthaMap (http://www.athamap.de/; Bülow et al., 2009) and then refined by referring to literature reports of the sequence features and functional properties of CREs (Supplemental Table S1). Because the promoter regions and first introns of the two genes contain most, if not all, of the CREs required for normal expression, special attention was paid to them.
To trace the evolutionary history of the TFBSs, we first obtained the genomic sequences of the AP1 and CAL homologs from four other brassicaceous species (Arabidopsis lyrata, C. rubella, Thellungiella parvula, and Aethionema arabicum) and two nonbrassicaceous species of the Brassicales (Tarenaya hassleriana of Cleomaceae and Carica papaya of Caricaceae) (Supplemental Table S3). The sequences of A. lyrata, C. rubella, and C. papaya were all obtained from Phytozome 9.1 (http://www.phytozome.net/) by TBLASTN searches. In the case of T. parvula, A. arabicum, and T. hassleriana, genome sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/). Exon-intron structures of these genes were annotated with Wise2 (http://www.ebi.ac.uk/Tools/psa/genewise/; Birney and Durbin, 2000), and TFBSs were determined based on sequence similarity and relative positions (Supplemental Table S2). Alignments of comparable regions were first generated in ClustalX 1.83 (Thompson et al., 1997) and then refined manually in GeneDoc (Nicholas et al., 1997). Phylogenetic relationships among the sampled species were determined based on the most recent study of the Brassicaceae (Couvreur et al., 2010). Gains and losses of TFBSs were inferred according to the maximum parsimony algorithm.
Expression Analysis
Total RNA was extracted from different tissues using the PureLink Plant RNA Reagent (Invitrogen) according to the user manual. First-strand complementary DNA (cDNA) was synthesized from 1 μg of total RNA using an oligo(dT) primer and the SuperScript III first-strand cDNA synthesis kit (Invitrogen), following the manufacturer’s instructions. qRT-PCR was performed using the PrimerScript RT Reagent Kit (Perfect Real Time; Takara) in the Applied Biosystems ViiA 7 Real-Time PCR System (Life Technologies). For each gene, at least two pairs of primers were designed, and their amplification efficiencies were determined by comparing the standard curves. Only primer pairs showing amplification efficiencies between 90% and 105% were used (Supplemental Table S4). Relative expression values were first normalized to a housekeeping gene, ACTIN, and then calculated by the comparative cycle threshold method (Livak and Schmittgen, 2001). All reactions were run in three biological replicates, each of which has three technical replicates. Statistical analyses of the qRT-PCR data were performed with the two-tailed Student’s t test.
For in situ hybridization, inflorescences with floral buds at various developmental stages were first fixed in 4% (w/t) paraformaldehyde and then embedded in Paraplast (Sigma). The 323-bp probe fragment specific to AP1 and the 313-bp probe fragment specific to CAL, both of which cover the C-terminal ends of their coding sequences, were amplified from cDNA using gene-specific primers, with the T7 adapter being introduced into the reverse primer (Supplemental Table S4). PCR products were used as templates for synthesizing antisense digoxigenin-labeled RNA probes with the DIG RNA Labeling Kit (Roche). Pretreatment, hybridization, and washing of sections (10 μm) were performed as described (Zhang et al., 2013), with minor modifications. The sections were exposed to 1 μg mL−1 proteinase K buffer for 30 min at 37°C before hybridization, and final washing of the hybridized sections was carried out in 0.5× SSC at 50°C for 30 min. Images were captured with a Zeiss Axio imager microscope.
Transgenic Constructs and Genetic Transformation
To generate the AP1pro2887+intr1:GUS construct, a 2,887-bp promoter fragment upstream of the translation start site and intron 1 of AP1 were amplified using the primer combinations AP1pro2887-F/AP1pro2887-R and AP1intr1-F/AP1intr1-R, respectively (Supplemental Table S4). Amplified fragments were cloned into the pEASY-Blunt Simple vector (TransGen Biotechnology) and assembled into the pENTR4 vector (Invitrogen) by digestion with appropriate restriction enzymes. The full-length fragment containing the promoter region and intron 1 of AP1 was subsequently transferred into pHGWFS7 destination vectors by LR Clonase reaction (Invitrogen). For the CALpro1210+intr1:GUS construct, a 1,210-bp promoter fragment and intron 1 of CAL were amplified with the primer combinations CALpro1210-F/CALpro1210-R and CALintr1-F/CALintr1-R, respectively (Supplemental Table S4). PCR-based mutagenesis was performed to yield the AP1pro2887(mut805-796)+intr1:GUS and CALpro1210(mut703-693)+intr1:GUS constructs (Supplemental Table S4). To generate constructs containing different lengths of AP1 or CAL promoters, truncated fragments were amplified from Col-0 genomic DNA by PCR using position-specific primers (Supplemental Table S4), and the amplified fragments were then recombined into the pHGWFS7 destination vector.
All recombinant plasmids were transferred into wild-type Arabidopsis (Col-0) plants using the Agrobacteriaum tumefaciens (GV3101)-mediated floral dip method (Clough and Bent, 1998). Seeds of transgenic plants were selected on solid one-half-strength Murashige and Skoog medium containing hygromycin (25 µg mL−1) and genotyped by PCR with GUS-specific primers (Supplemental Table S4). For each construct, at least 30 independent positive transgenic plants were analyzed in terms of GUS activity (Supplemental Table S5).
GUS Staining
Inflorescences under investigation were incubated in 90% (v/v) ice-cold acetone for 15 to 20 min, rinsed in the solution containing 100 mm sodium phosphate buffer (pH 7), 1 mm K3Fe(CN)6, and 1 mm K4Fe(CN)6, immersed into GUS staining solution containing 100 mm sodium phosphate buffer (pH 7), 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 10 mm EDTA, and 0.1% (v/v) Triton X-100, and then vacuum infiltrated until tissues became translucent. The materials were incubated overnight at 37°C in the GUS staining solution. After staining, the inflorescences were cleared with an ethanol series and maintained in 70% (v/v) ethanol. For anatomical observations, GUS-stained inflorescences were embedded and sectioned as described above. Whole-mount staining samples and histological sections were visualized using Leica S8 APO and Leica DM5000 B microscopes, respectively.
EMSA
Coding sequences of AP1, CAL, CruAP1, and CruCAL were amplified with the AthAP1-F/R, AthCAL-F/R, CruAP1-F/R, and CruCAL-F/R primer combinations, respectively (Supplemental Table S4). Amplified fragments were first digested and inserted into the pET28a expression vector (Novagen) and then transformed into Escherichia coli BL21(DE3) competent cells. Expression of the corresponding proteins was induced by adding 0.1 mm isopropyl β-d-thiogalactoside, and the concentration of proteins was measured with the BCA Protein Assay Kit (Pierce).
EMSA was done using the Light Shift Chemiluminescent EMSA Kit (Thermo Scientific). Briefly, 500 fmol of 5′-biotin-labeled probe DNA was incubated with 1.5 μg of poly(dI/dC) and 1 μg of proteins in 1× binding buffer at room temperature for 20 min. Reactions were then loaded onto a 6.5% (w/v) polyacrylamide gel (0.25× Tris-borate/EDTA) and run at 180 V constant for 1 h at 4°C. Blots were cross-linked using a Stratalinker-UV1800 device for 10 min.
Probe AP1 (5′-GACAAAATACTATTTTTGGGTTTGAAA-3′) was derived from the AP1 promoter. Probe CAL (5′-ATATTTCCTTAATTAACCCAAACTTC-3′) and probe CruAP1 (5′-ACAGAACACTTTTTCGGGTTTGAATAC-3′) were derived from the corresponding regions of probe AP1 in the CAL and CruAP1 promoters, respectively. The sequence of the positive control is 5′-AATACATTCCATATTTGGCAGGTGG-3′, which can be bound by AP1 in vitro (Huang et al., 1993; Riechmann et al., 1996b). The CArG box in probe AP1 and the positive probe is underlined.
ChIP-seq Data Analysis
A short sequence read data set from ChIP-seq experiments for AP1 (Kaufmann et al., 2010) was downloaded from the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/). Low-quality reads in the raw data were filtered out using FastQC software (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). The processed reads were then mapped to the Arabidopsis genome (The Arabidopsis Information Resource 10) using Bowtie2 (Langmead and Salzberg, 2012), allowing zero mismatches and only uniquely mapped reads to be counted. Peak calling was done using MACS2 (Zhang et al., 2008) with default parameters. ChIP-seq data were visualized using IGV (Thorvaldsdóttir et al., 2013).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Heat map showing the prevalence of expression divergence between duplicated MIKC-type MADS box genes in Arabidopsis.
Supplemental Figure S2. The DotPlot results of AP1 (6,946 bp, horizontal axis) and CAL (5,756 bp, vertical axis).
Supplemental Figure S3. Comparison of the promoter regions of AP1 and CAL.
Supplemental Figure S4. The DotPlot results of genomic sequences of AP1 (4,046 bp, horizontal axis) and CAL (3,756 bp, vertical axis).
Supplemental Figure S5. TFBS evolution.
Supplemental Figure S6. Alignments of representative TFBS-containing regions in a phylogenetic framework.
Supplemental Figure S7. CAL evolved under the less stringent constraint than AP1.
Supplemental Figure S8. Sequence alignment of ancestral and present-day AP1- and CAL-like proteins.
Supplemental Table S1. Functionally confirmed TFBSs in AP1 and CAL regulatory regions.
Supplemental Table S2. Sequences and positions of the TFBSs shown in Supplemental Figure S5.
Supplemental Table S3. AP1- and CAL-like genes included in this study.
Supplemental Table S4. Primer sequences used in this study.
Supplemental Table S5. Independent transgenic lines examined by GUS-staining analyses.
Supplementary Material
Acknowledgments
We thank Yalong Guo for providing C. rubella seeds; Xianxian Yu, Rui Zhang, and Peipei Wang for technical assistance in qRT-PCR and microscopy; Chunce Guo and Guixia Xu for help in bioinformatic and statistical analyses; and Hong Ma, Shin-Han Shiu, Kong laboratory members, and two anonymous reviewers for valuable comments.
Glossary
- CRE
cis-regulatory element
- TFBS
transcription factor-binding site
- qRT
quantitative real-time reverse transcription
- ChIP-seq
chromatin immunoprecipitation sequencing
- EMSA
electrophoretic mobility shift assay
- Col-0
Columbia-0
- cDNA
complementary DNA
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31125005 and 30800065), the Chinese Academy of Sciences Interdisciplinary Innovation Team, and the Chinese Academy of Sciences Youth Innovation Promotion Association.
Articles can be viewed without a subscription.
References
- Alvarez-Buylla ER, García-Ponce B, Garay-Arroyo A (2006) Unique and redundant functional domains of APETALA1 and CAULIFLOWER, two recently duplicated Arabidopsis thaliana floral MADS-box genes. J Exp Bot 57: 3099–3107 [DOI] [PubMed] [Google Scholar]
- Arthur W. (2011) Evolution: A Developmental Approach. Wiley-Blackwell Press, Oxford [Google Scholar]
- Baker CR, Hanson-Smith V, Johnson AD (2013) Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science 342: 104–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlloch R, Kim MC, Sayou C, Thévenon E, Parcy F, Nilsson O (2011) Integrating long-day flowering signals: a LEAFY binding site is essential for proper photoperiodic activation of APETALA1. Plant J 67: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Birney E, Durbin R (2000) Using GeneWise in the Drosophila annotation experiment. Genome Res 10: 547–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Bülow L, Engelmann S, Schindler M, Hehl R (2009) AthaMap, integrating transcriptional and post-transcriptional data. Nucleic Acids Res 37: D983–D986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Running MP, Williams RW, Meyerowitz EM (1999) The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev 13: 334–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC (2014) Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol 19: 91–98 [DOI] [PubMed] [Google Scholar]
- Couvreur TL, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K (2010) Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol Biol Evol 27: 55–71 [DOI] [PubMed] [Google Scholar]
- Crews ST, Pearson JC (2009) Transcriptional autoregulation in development. Curr Biol 19: R241–R246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M (1999) twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell 3: 297–307 [DOI] [PubMed] [Google Scholar]
- Davidson EH. (2006) The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Academic Press, San Diego [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES (2011) FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA 108: 6680–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R, Bergmiller T, Wagner A (2014) Increased gene dosage plays a predominant role in the initial stages of evolution of duplicate TEM-1 beta lactamase genes. Evolution 68: 1775–1791 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I (2012) Tempo and mode in evolution of transcriptional regulation. PLoS Genet 8: e1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Andrés F, Sessa A, Guerra RF, Simonini S, Mateos JL, Torti S, Zambelli F, Prazzoli GM, Bjerkan KN, et al. (2013) Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis. Genome Biol 14: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson-Brown C, Savidge B, Yanofsky MF (1994) Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76: 131–143 [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang C, Yang H, Jiao Y (2014) Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Natl Acad Sci USA 111: 6840–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Taylor J (2012) Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet 13: 469–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J (2005) Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Mizukami Y, Hu Y, Ma H (1993) Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res 21: 4769–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tudor M, Su T, Zhang Y, Hu Y, Ma H (1996) DNA binding properties of two Arabidopsis MADS domain proteins: binding consensus and dimer formation. Plant Cell 8: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Klein J, Lönnig WE, Meijer H, Saedler H, Sommer H (1992) Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J 11: 1239–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Posé D, Ferrario S, Ott F, Kaufmann K, Valentim FL, de Folter S, van der Wal F, van Dijk AD, Schmid M, et al. (2012) Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol 160: 433–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Levy M, Pilpel Y (2006) The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci USA 103: 11653–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Anfang N, Saedler H, Theissen G (2005) Mutant analysis, protein-protein interactions and subcellular localization of the Arabidopsis B sister (ABS) protein. Mol Genet Genomics 274: 103–118 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. (2010) Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267: 522–525 [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Bancewicz RM, Gautier P, Dahm R, Schonthaler HB, Damante G, Seawright A, Hever AM, Yeyati PL, van Heyningen V, et al. (2008) Subfunctionalization of duplicated zebrafish pax6 genes by cis-regulatory divergence. PLoS Genet 4: e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton-Rauh AL, Buckler ES IV, Purugganan MD (1999) Patterns of molecular evolution among paralogous floral homeotic genes. Mol Biol Evol 16: 1037–1045 [DOI] [PubMed] [Google Scholar]
- Lenser T, Theissen G, Dittrich P (2009) Developmental robustness by obligate interaction of class B floral homeotic genes and proteins. PLOS Comput Biol 5: e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Yang J, Gu X (2005) Expression divergence between duplicate genes. Trends Genet 21: 602–607 [DOI] [PubMed] [Google Scholar]
- Litt A. (2007) An evaluation of A-function: evidence from the APETALA1 and APETALA2 gene lineages. Int J Plant Sci 168: 73–91 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-Δ Δ CT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: analysis and visualization of genetic variation. Embnew News 4: 14 [Google Scholar]
- Ohno S. (1970) Evolution by Gene Duplication. Springer, New York [Google Scholar]
- Ó’Maoiléidigh DS, Wuest SE, Rae L, Raganelli A, Ryan PT, Kwasniewska K, Das P, Lohan AJ, Loftus B, Graciet E, et al. (2013) Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D (1998) A genetic framework for floral patterning. Nature 395: 561–566 [DOI] [PubMed] [Google Scholar]
- Pastore JJ, Limpuangthip A, Yamaguchi N, Wu MF, Sang Y, Han SK, Malaspina L, Chavdaroff N, Yamaguchi A, Wagner D (2011) LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development 138: 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Posé D, Verhage L, Ott F, Yant L, Mathieu J, Angenent GC, Immink RG, Schmid M (2013) Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417 [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Carroll SB (2007) Emerging principles of regulatory evolution. Proc Natl Acad Sci USA (Suppl 1) 104: 8605–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Krizek BA, Meyerowitz EM (1996a) Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc Natl Acad Sci USA 93: 4793–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang M, Meyerowitz EM (1996b) DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res 24: 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB. (2014) Complexity of gene expression evolution after duplication: protein dosage rebalancing. Genet Res Int 2014: 516508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y (2012) Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet 13: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D (2006) The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development 133: 1673–1682 [DOI] [PubMed] [Google Scholar]
- Schauer SE, Schlüter PM, Baskar R, Gheyselinck J, Bolaños A, Curtis MD, Grossniklaus U (2009) Intronic regulatory elements determine the divergent expression patterns of AGAMOUS-LIKE6 subfamily members in Arabidopsis. Plant J 59: 987–1000 [DOI] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH (2007) Consequences of genome duplication. Curr Opin Genet Dev 17: 505–512 [DOI] [PubMed] [Google Scholar]
- Shan H, Zhang N, Liu C, Xu G, Zhang J, Chen Z, Kong H (2007) Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol Phylogenet Evol 44: 26–41 [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. (2012) Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R (1998) Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development 125: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Sundström JF, Nakayama N, Glimelius K, Irish VF (2006) Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis. Plant J 46: 593–600 [DOI] [PubMed] [Google Scholar]
- Taylor SA, Hofer JM, Murfet IC, Sollinger JD, Singer SR, Knox MR, Ellis TH (2002) PROLIFERATING INFLORESCENCE MERISTEM, a MADS-box gene that regulates floral meristem identity in pea. Plant Physiol 129: 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM (2004) Gene regulatory network growth by duplication. Nat Genet 36: 492–496 [DOI] [PubMed] [Google Scholar]
- Theissen G. (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4: 75–85 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Flicek P, Odom DT (2014) Evolution of transcription factor binding in metazoans: mechanisms and functional implications. Nat Rev Genet 15: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584 [DOI] [PubMed] [Google Scholar]
- Wang B, Zhang N, Guo CC, Xu GX, Kong HZ, Shan HY (2012) Evolutionary divergence of the APETALA1 and CAULIFLOWER proteins. J Syst Evol 50: 502–511 [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D (2004) Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA 101: 1775–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13: 59–69 [DOI] [PubMed] [Google Scholar]
- Wray GA. (2007) The evolutionary significance of cis-regulatory mutations. Nat Rev Genet 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419 [DOI] [PubMed] [Google Scholar]
- Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F (2012) Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA 109: 13452–13457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, McKim SM, Murmu J, Haughn GW, Hepworth SR (2010) Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J 63: 974–989 [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M (2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E (2011) AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res 39: D1118–D1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Guo C, Zhang W, Wang P, Li L, Duan X, Du Q, Zhao L, Shan H, Hodges SA, et al. (2013) Disruption of the petal identity gene APETALA3-3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proc Natl Acad Sci USA 110: 5074–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.