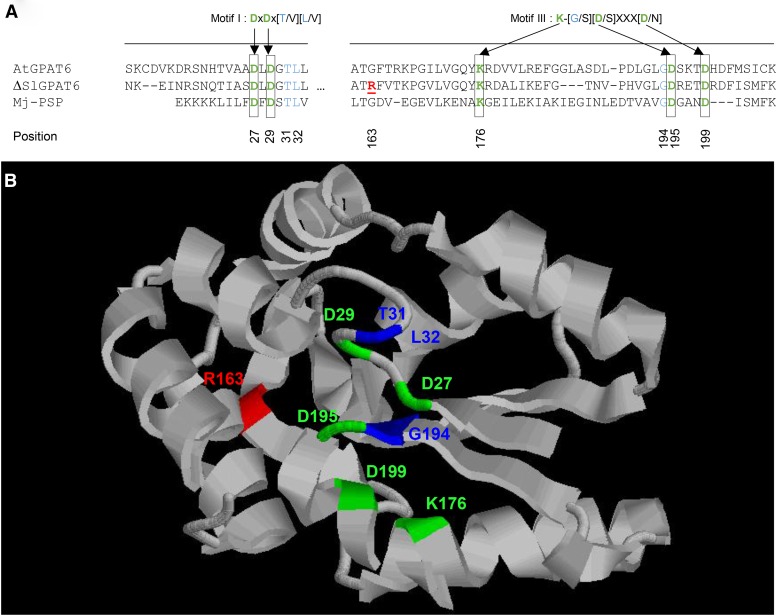
Figure 3.
Mutation positions in the ΔSlGPAT6 protein. A, Protein sequence alignment of Arabidopsis AtGPAT6, the tomato ΔSlGPAT6 variant, and the Methanococcus jannaschii phospho-Ser phosphatase (Mj-PSP). Black boxes show residues required for PSP activity in motifs I and III, according to Yang et al. (2010). The G163R mutation is indicated as underlined boldface red text. B, In silico ΔGPAT6 modeling based on Protein Data Bank structure 1L7P, restricted to amino acid positions 17 to 207. Critical residues from motifs I and III are indicated in green for D/K amino acids and in blue for other amino acids from canonical motifs. The position of the mutation is indicated in red.