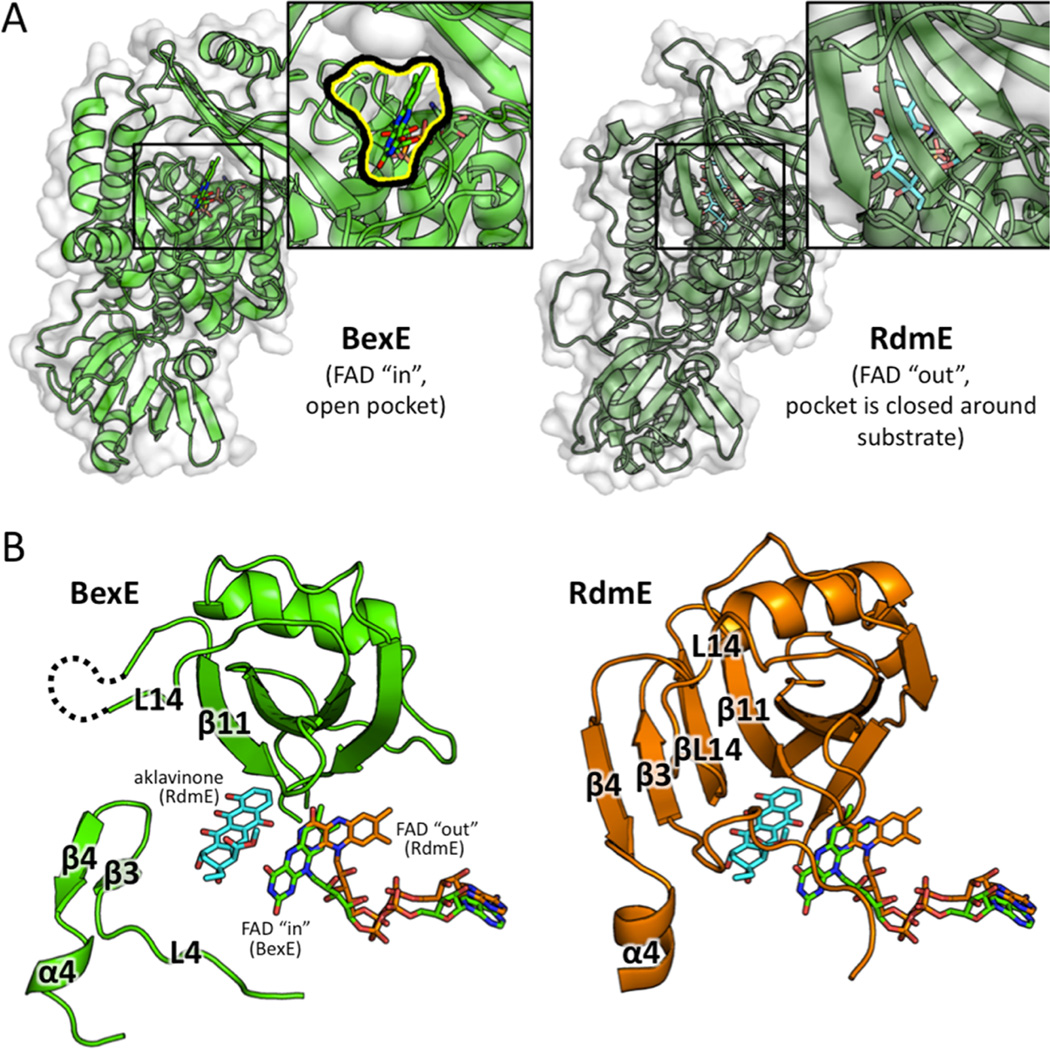
Figure 6.
A structural comparison between the middle domains of BexE with FAD in the “in” position and RdmE bound to aklavinone and FAD in the “out” position. (A) BexE with FAD in the “in” position with a zoomed in view of the putative active site entrance (left). RdmE bound to aklavinone with FAD in the “out” position with a zoomed in view of the area corresponding to the BexE active site entrance (right). In RdmE, there is no active site entrance in this area where the substrate is bound. (B) In BexE (left, light green), FAD is in the “in” position with an open active site. The β-hairpin consisting of β3 and β4 is relaxed and shifted downward compared to the same region in RdmE (right, dark green). The L14 region of BexE could not be modeled because of lack of electron density; however, in RdmE this loop forms a β-strand (βL14), which forms a β-sheet with β3, β4, and β11. *Aklavinone, and FAD from both structures are overlaid in both BexE and RdmE to highlight the FAD motion with respect to substrate position.