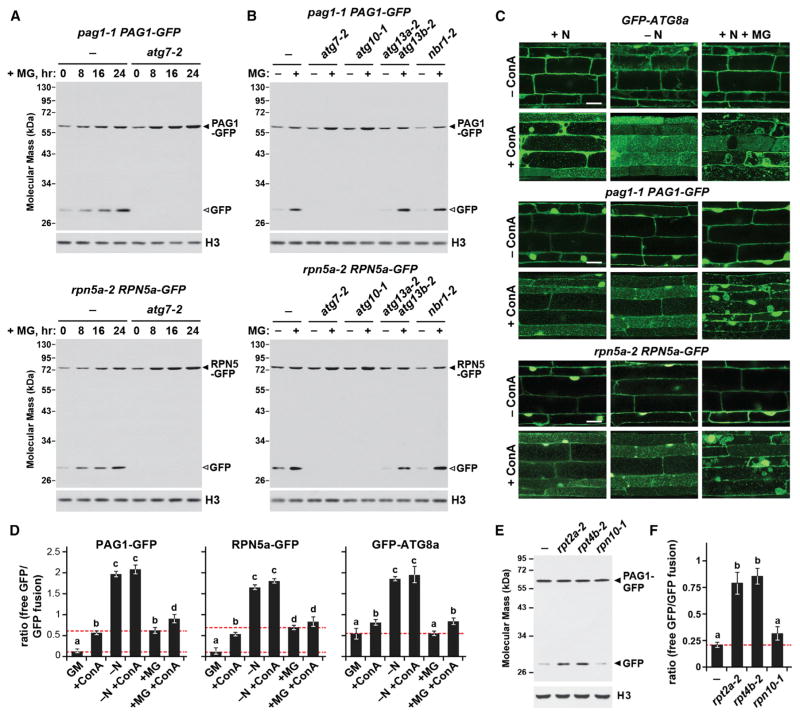
Figure 3. Proteaphagy Is Stimulated by Chemical or Genetic Inhibition of the Proteasome.
(A) Time course of free GFP release from PAG1-GFP and RPN5a-GFP following MG132 exposure. WT and atg7-2 seedlings were exposed to 50 μM MG132 (MG) for the indicated times, and total protein extracts were immunoblotted with anti-GFP antibodies. The GFP fusions and free GFP are indicated by closed and open arrowheads, respectively.
(B) Effects of various autophagy mutants on the cleavage of PAG1-GFP and RPN5a-GFP during MG132 exposure. WT, atg7-2, atg10-1, atg13a-2 atg13b-2, and nbr1-2 seedlings were incubated with or without 50 μM MG132 for 16 hr. Immunoblotting was performed as in (A).
(C) MG132 induces the delivery of autophagic vesicles to the vacuole. WT seedlings were either N-fed or starved and/or exposed to 50 μM MG132 and/or 1 μM ConA for 16 hr. Root cells were examined for autophagic bodies by confocal microscopy. Scale bar, 10 μm.
(D) Quantification of the free GFP/tagged-GFP ratio during N starvation or MG132 treatment of the seedlings analyzed in (C). Corresponding immunoblots are shown in Figure S6C. Bars represent the mean (± SD) of three biological replicates. Letters indicate values that are statistically different from one another (p < 0.05).
(E) The proteasome mutants rpt2a-2 and rpt4b-2, but not rpn10-1, stimulate proteaphagy. Well fed seedlings were assayed for the release of free GFP by immunoblotting as in (A).
(F) Quantification of the free GFP/PAG1-GFP ratio in the proteasome mutants by densitometric scans of the immunoblot shown in (E). Bars represent the mean (± SD) of three biological replicates. Letters indicate values that are statistically different from one another (p < 0.05).
See also Figures S3–S6.