Abstract
We introduce a melded chemical and computational approach for probing and modeling higher-order intramolecular tertiary interactions in RNA. 2'-Hydroxyl molecular interference (HMX) identifies nucleotides in highly packed regions of an RNA by exploiting the ability of bulky adducts at the 2'-hydroxyl position to disrupt overall RNA structure. HMX was found to be exceptionally selective for quantitative detection of higher-order and tertiary interactions. When incorporated as experimental constraints in discrete molecular dynamics (DMD) simulations, HMX information yielded accurate three-dimensional models, emphasizing the power of molecular interference to guide RNA tertiary structure analysis and fold refinement. In the case of a large, multi-domain RNA, the Tetrahymena group I intron, HMX identified multiple distinct sets of tertiary structure interaction groups in a single, concise experiment.
Graphical abstract
Introduction
RNA plays diverse and central roles in the regulation of gene expression.1 Information is encoded in the RNA at several levels: the primary sequence, the specific base-pairing pattern that defines the secondary structure, and higher-order RNA structures composed of tightly packed secondary structure elements stabilized by a few key tertiary interactions.2 The precise formation of higher-order tertiary structures is critical to the function of many RNAs.3,4 RNA secondary and tertiary interactions can be interrogated by modifying an RNA with chemical probes or by incorporating nucleotide substitutions that disrupt native structure. In modification interference, an RNA is treated to introduce chemical modifications, usually at the nucleobases, and then the RNA is subjected to a partitioning experiment to distinguish functional from non-functional molecules.5–7 For the nucleotide analog interference mapping (NAIM)8,9 strategy, nucleotide analogs are incorporated into an RNA transcript, and active RNAs are partitioned from those that are inactivated due to the nucleotide analog. Both modification interference and NAIM can interrogate most nucleotides in an RNA to identify single nucleotide or single atom interactions, respectively, critical to the tertiary structure.5,6,8,9 These approaches generally require multiple distinct experiments to interrogate the tertiary environment of every nucleotide in an RNA.
Chemical probes are also widely used to examine both solvent accessibility and dynamics in the RNA backbone. The solvent accessibility of the RNA backbone can be monitored by hydroxyl radical footprinting (HRP).10,11 Backbone dynamics can be monitored in the selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) strategy that uses reagents sensitive to the nucleophilicity of the 2'-OH group, which is dependent on the underlying flexibility of the nucleotide.12–14 Reactivities of these and other chemical probes, like DMS, CMCT and kethoxal, are modulated by both secondary and tertiary structure interactions, and it is usually difficult to deconvolute the relative influence of each type of interaction.
Here, we describe a strategy in which 2'-hydroxyl-selective reagents are used in a modification-interference experiment to simply, directly and specifically interrogate RNA tertiary structure. In this approach, which we call 2'-hydroxyl molecular interference, or HMX, a hydroxyl-selective reagent is used to create a pool of RNAs with evenly distributed 2'-O-ester adducts. Next, a structure-selective pressure, such as RNA folding, is placed on the pool of modified RNAs. A subset of 2'-O-ester groups will interfere with molecular interactions and prevent native structure formation. By partitioning the sample into folded and unfolded states, nucleotides whose modification disrupts tertiary interactions are identified. Here we use this information to characterize the internal packing interactions that define higher-order RNA structure, to refine three-dimensional structure models, and to detect multiple sets of tertiary interactions in a large complexly folded RNA.
Results
HMX overview
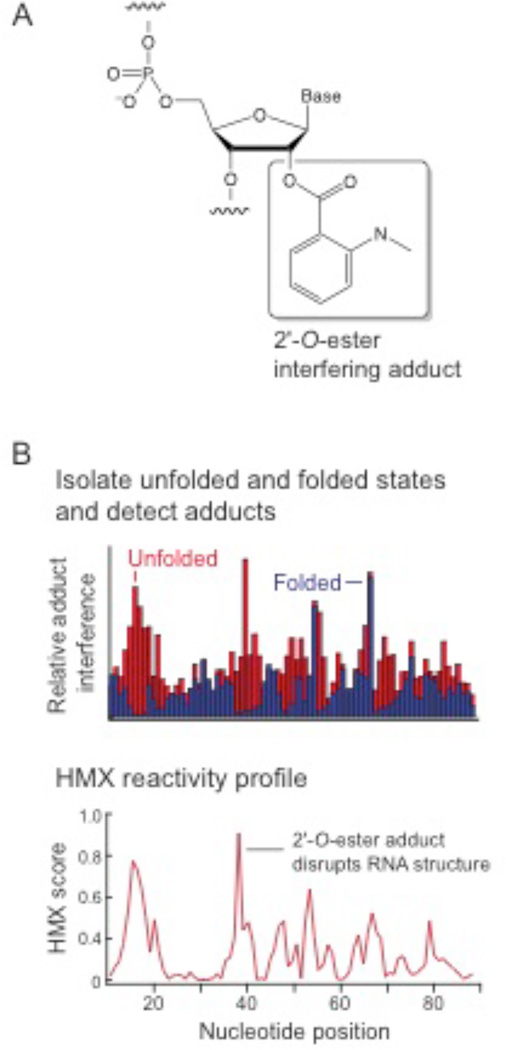
In the first step of the HMX strategy, an RNA of interest is modified with a 2’-hydroxyl selective reagent under denaturing conditions such that modifications are distributed roughly equally and sparsely among all nucleotides in the RNA population. Second, the RNA is allowed to fold under conditions that favor the native, functional state; and, third, the RNA is subjected to a selection step to partition the RNA into active and inactive components. An experiment with an unmodified control is performed in parallel. The RNAs in this analysis were modified using N-methylisatoic anhydride (NMIA).12 NMIA is particularly well-suited for this application because it modifies RNA at high temperatures, to form a simple bulky (but not too bulky) adduct in the RNA backbone. Under denaturing conditions (95 °C at low ion concentrations) NMIA modifies all positions in an RNA at the 2'-hydroxyl position (Fig. 1A and Supp. Fig. 1). Some adducts will have no or small structural consequences, whereas other adducts will prevent native folding of the RNA. We partitioned the natively folded from the unfolded structures based on mobility in non-denaturing acrylamide gels, although many other selection strategies are compatible with this approach. After partitioning folded and unfolded populations, positions of modified nucleotides were detected as stops using reverse transcription-mediated primer extension. Adducts that disrupted folding were identified by comparing the profiles of the unfolded and folded RNA at each position (Fig. 1B). HMX scores for each nucleotide were calculated as the difference between the normalized profiles for the folded and unfolded RNA.
Figure 1.
2'-Hydroxyl molecular interference (HMX). (A) Structure of the 2'-O-ester adduct introduced by reacting RNA with NMIA. Select 2'-hydroxyl adducts interfere with native folding, creating a population of unfolded RNA that can be partitioned from fully folded RNA. (B) Partitioned populations are separately analyzed for function-disrupting chemical adducts. HMX profiles are calculated by cross-correlation.
We initially explored the HMX approach using structurally diverse RNAs for which high-resolution structures are available: yeast tRNAAsp (75 nts),15 E. coli thiamine pyrophosphate (TPP) riboswitch (79 nts),16 the B. subtilis M-Box riboswitch (156 nucleotides),17 and the Tetrahymena group I intron P546 domain (160 nts).18 After modifying the RNA with NMIA under denaturing conditions, RNAs were folded under conditions that stabilize their native and tertiary structure. The TPP and M-Box riboswitch RNAs were folded in the presence of saturating ligand concentrations.
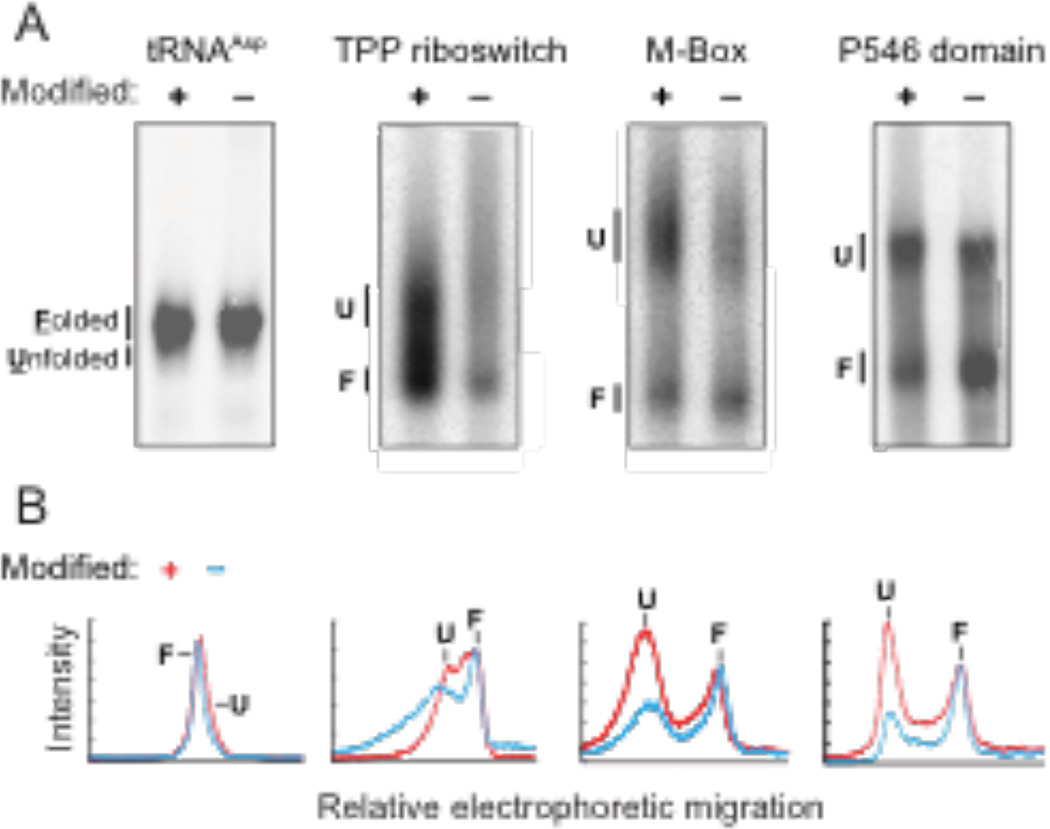
The unfolded RNA state was identified by an increase in population of a species in the modified RNA as compared to the unmodified RNA (Fig. 2A). In the case of the TPP and M-Box riboswitches, and the P546 domain, there was a clear shift to a second (unfolded and less compact) state in the modified RNA, relative to the unmodified control RNA (Fig. 2B). In the case of tRNAAsp, folded and unfolded separated, but just barely. The unfolded state for tRNAAsp migrated slightly more rapidly than the folded state, implying that the L-shaped tertiary structure causes the folded RNA to migrate more slowly than does the unfolded structure, consistent with the known behavior of bent RNAs.19
Figure 2.
Partitioning of RNA populations. (A) Folded and unfolded populations for modified and unmodified RNAs separated by non-denaturing gel electrophoresis in the presence of 50 mM NaCl and 5 mM MgCl2. For clarity, gel images were straightened and scaled to show similar representations for each RNA; band intensity profiles are unaltered. (B) Band intensities as a function of gel migration distance.
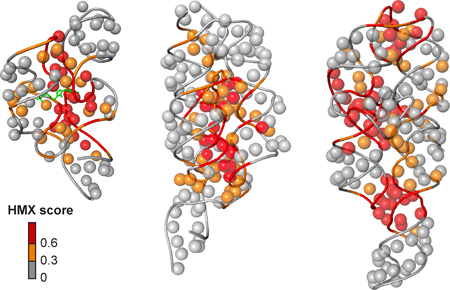
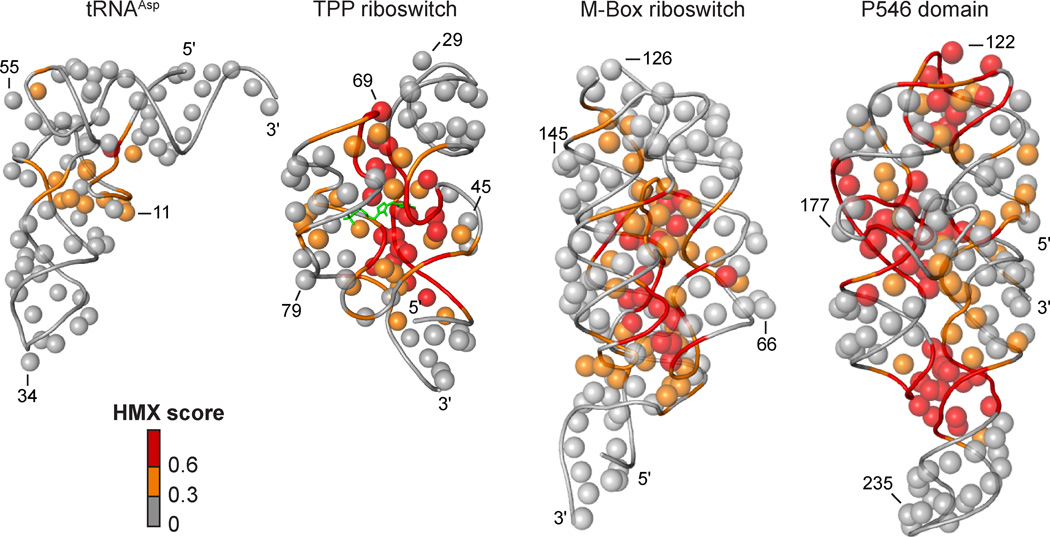
After partitioning, sites of modification were identified in the folded and unfolded populations by reverse transcription-mediated primer extension. 2'-O-ester adducts that prevent folding were over represented in the unfolded band and underrepresented in the folded band (Supp. Fig. 2A). The resulting modified RNA data were normalized using a cross-correlation approach to create an HMX score that allowed identification of nucleotides preferentially modified in the unfolded population relative to the folded population. The HMX score takes into account that the separation of unfolded and folded populations using 2'-O-adduct molecular interference is imperfect and that there is some noise in the separated signals (Methods and Supp. Fig. 2). Positions with medium and high interference scores were visualized on the known three-dimensional structures15–18 of each RNA (Fig. 3). Nucleotides with high HMX scores corresponded to nucleotides directly involved in tertiary interactions and to nucleotides within densely packed regions of the RNA. Because the 2'-O-ribose modification occurs in the RNA backbone and likely does not significantly destabilize helix formation,20,21 interfering positions corresponded almost exclusively to higher-order interactions and not to canonically base-paired nucleotides (Fig. 3).
Figure 3.
Visualization of HMX interference information on accepted three-dimensional structures.15–18 The 2'-OH group for each nucleotide is shown as a sphere and the phosphoribose backbone by a tube. Nucleotides are colored by HMX score; the TPP ligand is green.
Molecular overlap model for HMX intensities
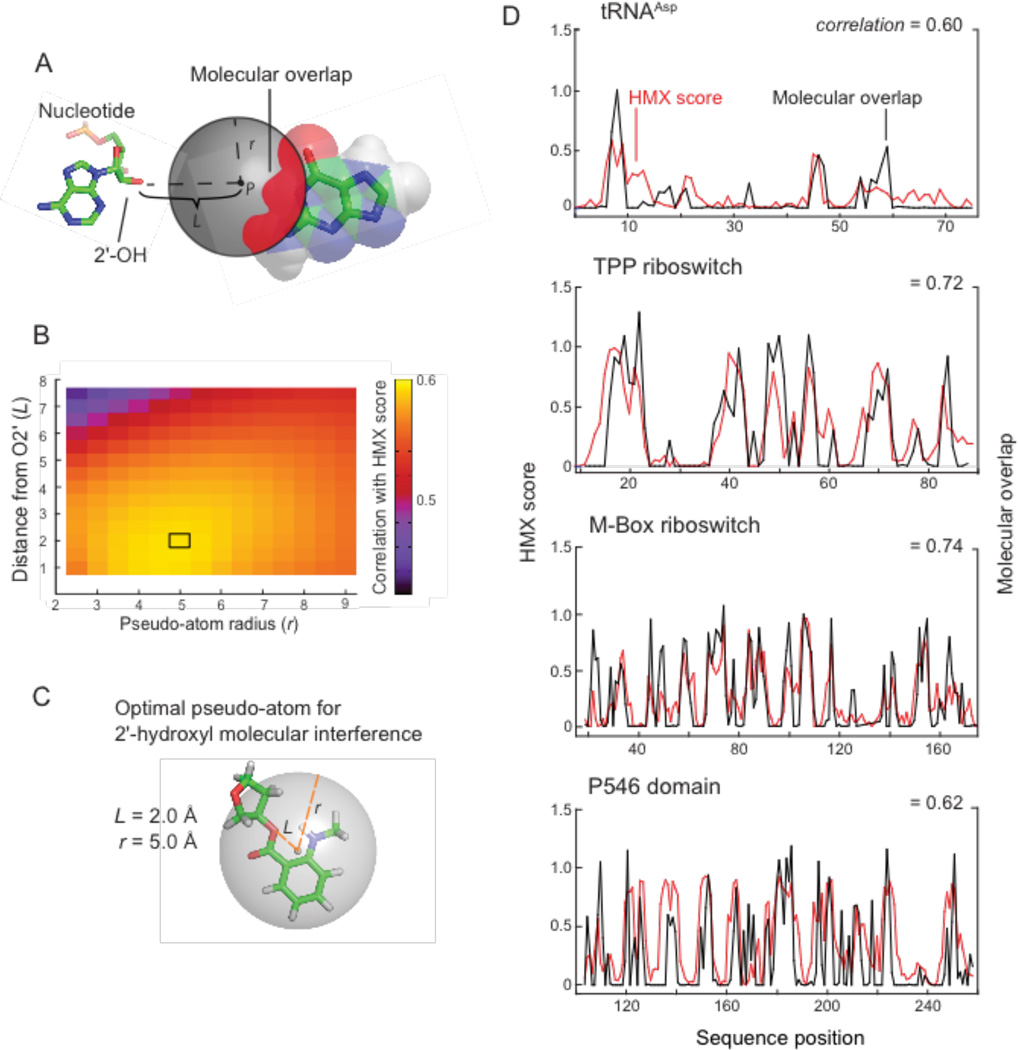
Because molecular interference appeared to correlate so strongly with RNA tertiary interactions, we sought to understand the molecular basis of this correlation. To do so we first defined a pseudo-atom, representing the 2'-O-ester adduct, described by two parameters: L, the length of the pseudo-atom vector extended from the 2'-carbon–2'-oxygen bond, and r, the radius of the pseudo-atom. Using the accepted three-dimensional RNA structures we calculated the degree to which surrounding nucleotide atoms intersected the defined pseudo-atom shell, based on their van der Waals radii (Fig. 4A). The pseudo-atom bond length and atomic radius were determined by calculating a correlation coefficient between the simulated and the experimental interference score (Fig. 4B). The pseudoatom parameters that best fit the experimental data for all RNAs were L of 2 Å and r of 5 Å. A pseudo-atom with these parameters tightly, and fully, encapsulates the NMIA adduct ester at a ribose ring (Fig. 4C).
Figure 4.
Physical model for 2'-hydroxyl molecular interference. (A) Model for interference by molecular overlap in which adducts are represented by a pseudo-atom (grey) at a distance (L) from the O2' position at radius (r). (B) Analysis of optimal pseudo-atom bond length and atomic radius. Maximum correlation between Pearson's r and pseudo-atom representing 2'-hydroxyl molecular interference is boxed. (C) Relationship between pseudo-atom dimensions and 2'-O-ester adduct. (D) Relationship between HMX scores and molecular overlap for tRNAAsp, the TPP and M-Box riboswitches, and P546 domain RNAs. HMX score profiles (red) show a high correlation with calculated molecular overlaps (black) for each RNA. Pearson correlation coefficients are shown.
The correlations between the experimental interference scores and the molecular overlap calculations for each RNA are high (Fig. 4D), indicating that the 2'-O-ester adduct disrupts RNA structure by sterically blocking RNA interactions in crowded regions of the RNA. For example, interfering nucleotides in the TPP riboswitch interact directly with ligand and other nucleotides form RNA-RNA contacts. The HMX experiment was sensitive to both types of interactions, indicating that HMX will be useful for examining intramolecular and intermolecular RNA contacts and protein and small molecule ligand interactions with RNA. Critically, as judged by visualizing interfering positions in three dimensions (Fig. 3) and from molecular overlap analysis (Fig. 4D), HMX analysis is exquisitely sensitive to higher-order molecular interactions in RNA, with essentially no detection of false positive interactions.
Three-dimensional RNA structure modeling
The HMX strategy yields a quantitative physical measurement related to the extent of RNA tertiary structure packing at the site of an individual nucleotide. We therefore explored whether HMX scores could be used to refine three-dimensional models for RNA. Our labs have previously shown that hydroxyl radical probing (HRP) data, which are roughly related to solvent accessibility, can be used as structural constraints in discrete molecular dynamics simulations22,23 to increase the quality of structural models relative to unconstrained simulations.24 Implementation of the HRP-based potential is experimentally and computationally difficult, because HRP data are inherently noisy, especially for smaller RNAs with few solvent-protected nucleotides. We adapted the procedure previously developed for HRP-refined structures24,25 to develop three-dimensional models for these four RNAs based on HMX constraints. DMD simulations were performed using a simplified model in which each nucleotide was represented by three pseudo-atoms corresponding to the base, sugar, and phosphate groups.23 The energy function used to direct folding included terms for both bonded and non-bonded interactions. Additional potential energy terms incorporated information on the number of tertiary contacts as estimated from HMX data. Structures obtained by simulation were clustered based on structural similarity to generate a representative model for each RNA.
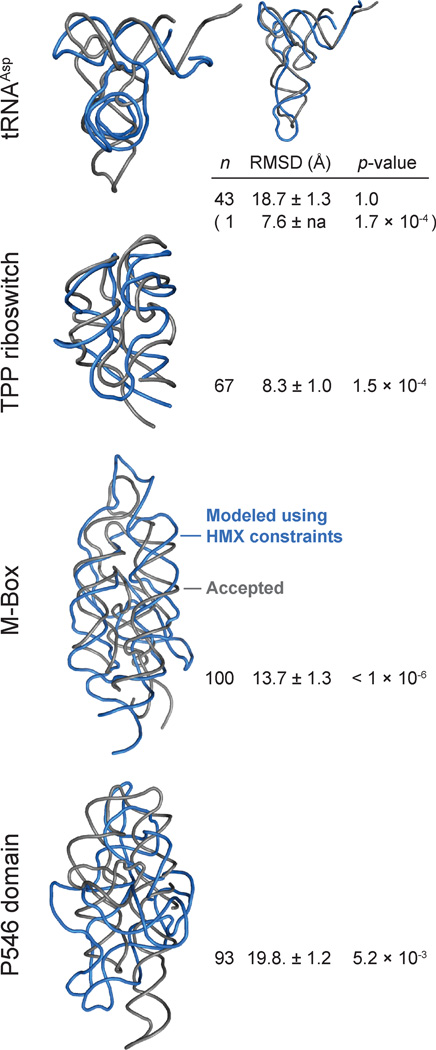
The predicted structural models for each RNA were evaluated in terms of number and population of clusters as well as the calculated root-mean-square deviation (RMSD) from the accepted crystal structures. HMX-directed simulations produced structurally and statistically significant26 native-like RNA structured models for the TPP and M-Box riboswitches and for the P546 domain (Fig. 5). For each RNA, the central structure in the highest population cluster contained the structure with the lowest RMSD relative to the accepted structure. In contrast, HMX-directed simulations for tRNAAsp did not produce a single structure and the model with the lowest RMSD corresponded to the lowest population cluster (Fig. 5; top). Critically, the tRNAAsp simulation itself revealed that no well-determined model emerged, as no single cluster included a majority of structures consistent with the molecular interference information (Fig. 5 and Table S1). Thus, the HMX-directed simulations proved highly robust, did not return a false-positive model when data were insufficient (as in the case of tRNAAsp), and yielded highly significant models for RNAs as large as 160 nts.
Figure 5.
HMX-directed RNA fold refinements. RNAs are shown as backbone traces. Accepted structures15–18 and HMX-directed refinements for each RNA are gray and blue, respectively. The cluster populations (n), mean RMSD, and p-values26 are shown. For tRNAAsp, both the largest cluster (large image) and lowest RMSD structures (inset) are shown.
HMX reveals individual structural domains in a large group I intron RNA
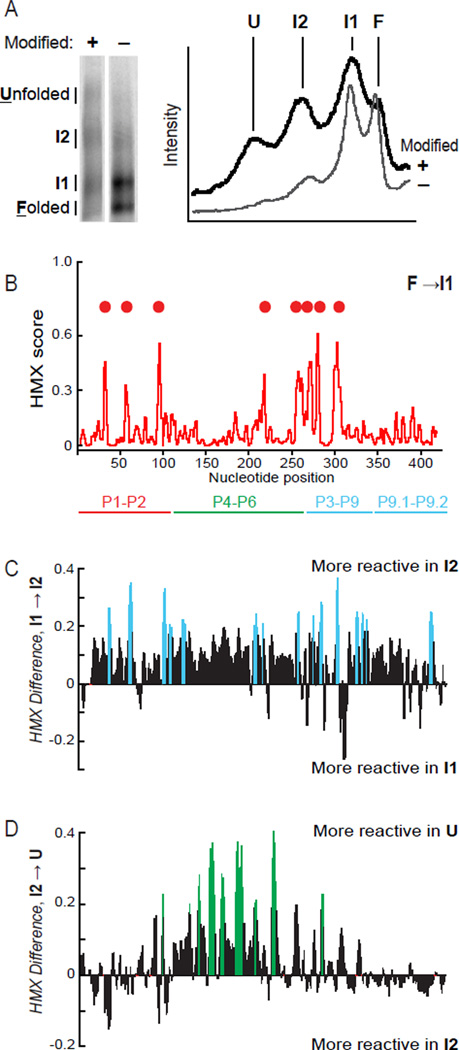
The Tetrahymena group I intron is formed from a set of conserved RNA domains. These domains include the P5-P4-P6 and P9-P7-P3-P8 domains, which form the active site of the RNA, and a P1–P2 helix that contains the 5' splice site.27,28 When the Tetrahymena group I intron RNA construct (~420 nts), designed to recapitulate the intron structure visualized by crystallography,29 was subjected to the HMX experiment, partitioning immediately revealed multiple unfolded states (Fig. 6A). The fastest migrating band is the most native-like structure and each subsequent lower-mobility band represents an RNA state with decreasing levels of tertiary structure, with the slowest migrating band reflected an RNA with no tertiary structure. HMX scores for each unfolded band were calculated by comparing each state to the fully folded RNA.
Figure 6.
HMX analysis of the Tetrahymena group I intron. (A) Partitioning of the RNA reveals folded and unfolded states, plus two intermediate states, labeled I1 and I2. Gel images (left) were straightened; gel lane intensity profiles (right) are unmanipulated. (B) HMX profile for the first (I1) intermediate, relative to the fully folded band. Sites of strongest interference are indicated with spheres. Difference HMX profiles calculated from the HMX profiles for the I1, I2, and unfolded bands (Supp. Fig. 3), correspond to transitions (C) from the first (I1) to the second (I2) intermediates, and (D) from I2 to the fully unfolded RNA. Differences larger than 0.2 HMX units are emphasized in color.
The HMX profile for the first structural intermediate, I1, reveals eight regions with high HMX scores (Fig. 6B). These positions fall primarily in the P1-P2 helices and in the P9-P7-P3-P8 domain around the intron active site (Fig. 6B, red circles). The first intermediate band thus corresponds to a structure in which 2'-hydroxyl interference disrupts docking of the P1-P2 helix via interactions with the P9-P7-P3-P8 domain (Fig. 7, in red).
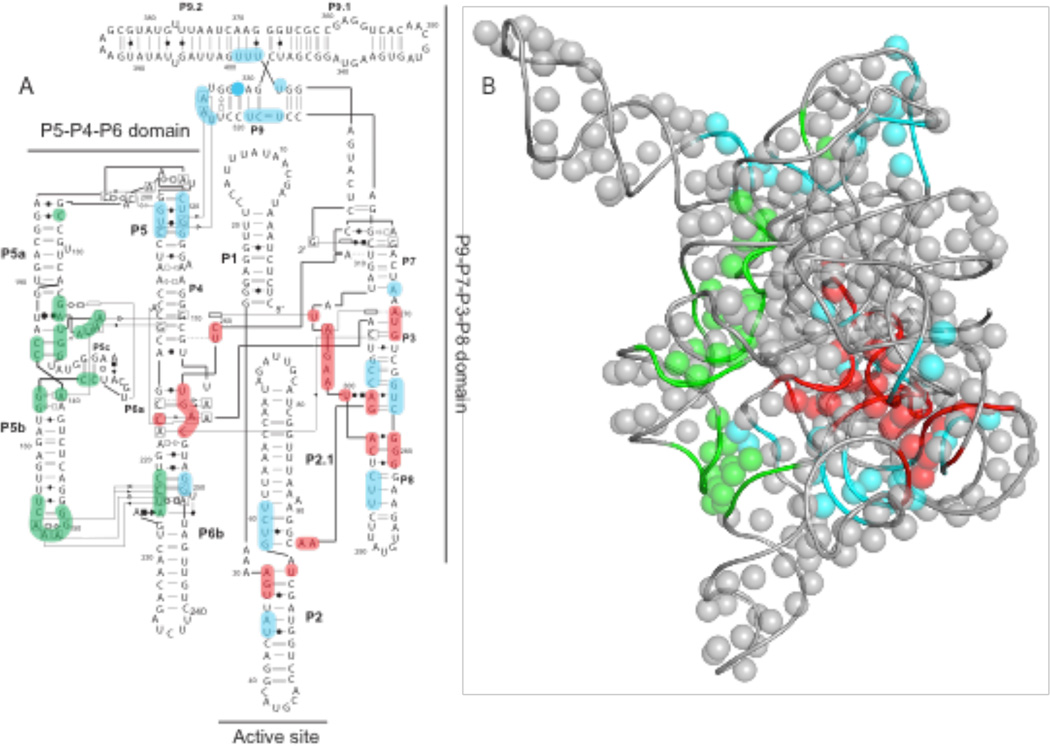
Figure 7.
Visualization of 2'-O-ester interferences on the (A) secondary and (B) tertiary structure of the Tetrahymena group I intron RNA. Unique interferences corresponding to the I1, I2 and fully unfolded RNA states (see Fig. 6) are colored red, blue and green, respectively. In panel B, 2'-OH groups are shown with spheres and the phosphoribose backbone as a tube. The three-dimensional structure corresponds to an extended model32 based on the crystal structure of core regions.29
To visualize the specific nucleotides whose modification induced formation of the I2 and fully unfolded states, we calculated difference HMX scores for these two transition (Fig. 6C,D and Supp. Fig. 3). Analysis of differences between I1 and the second intermediate, I2, reveals a large number of newly interfering nucleotides, involving many of the long-range tertiary contacts that stabilize the interaction between the P9-P7-P3-P8 and P5-P4-P6 domains. (Fig. 6C and 7A, in blue). Changes in interference profiles between I2 and the fully unfolded RNA fall almost exclusively in the P5-P4-P6 domain (Fig. 6D and 7A, in green). Taken together, these data indicate that the I2 intermediate corresponds to a state in which most or all tertiary interactions have been disrupted except those in the P5-P4-P6 domain. The transition from the I2 intermediate to the fully unfolded state then reports unfolding of this latter domain and loss of all remaining tertiary interactions.
This single HMX experiment (Fig. 6) reveals a hierarchy of tertiary interactions fully consistent with extensive prior crystallographic and biochemical analyses27,28 Moreover, HMX also emphasizes the high degrees of cooperativity in these interactions, as 2'-O-ester interference in any of the three structural networks of interfering nucleotides yield similar folding intermediate states (Fig. 7).
Discussion
HMX measures the effect of introducing a molecular perturbation at the ribose 2'-OH position on RNA folding. Modifications at the 2'-ribose position, which lie on the exterior of an RNA duplex, generally do not substantially destabilize simple RNA secondary structures20,21. Thus, the 2'-O-ester molecular interference measurement is exquisitely and specifically sensitive to interactions that govern RNA tertiary folding. For the five RNA evaluated in this work – tRNAAsp, the TPP and M-Box riboswitch aptamer domains, the P546 domain RNA, and the intact Tetrahymena group I intron – the interfering nucleotides identified by HMX correspond closely to the densely packed interior of these structures (Figs. 3 and 7B). This relationship is quantitative. Molecular interference by the 2'-O-ester group was highly correlated with a sphere of defined location relative to the RNA ribose group (Fig. 4). We anticipate that 2'-O-ester mediated molecular interference will prove broadly useful in evaluating higher-order RNA packing in the context of large RNAs and RNA-protein complexes.
The gel electrophoresis partitioning approach used here revealed the high level of cooperativity and structural interdependency in RNA tertiary folding. For the TPP and M-Box riboswitch aptamer domains and the P546 domain RNA, the introduction of distinct molecular adducts resulted in a single, well-defined, predominant unfolded structural population (Fig. 2), rather than multiple populations that would be expected due to the absence of individual tertiary interactions. There thus exists a high degree of cooperativity in the folding of these RNAs of up to 160 nucleotides.
Use of HMX information to direct a DMD-based three-dimensional structure refinement yielded highly significant structure models, relative to accepted RNA structures. Refinement was based on a recently developed approach that allows solvent accessibility to be incorporated into the simulation as an energy restraint.24 In general, use of molecular interference information required fewer assumptions be introduced and resulted in final clustered structures that were better defined than those obtained with hydroxyl radical restraints. For the TPP and M-Box riboswitches and the P546 domain RNAs, HMX-directed refinement yielded a single predominant cluster with highly statistically significant agreement to the accepted structure (Fig. 5). tRNAAsp was poorly modeled, likely reflecting both the inability to completely separate folded and unfolded states (Fig. 2) and the resulting low magnitude molecular interference data (Fig. 3, compare tRNA with other RNAs). Critically, the simulation itself clearly reported that this RNA was not a good target for refinement because a relatively large number of clusters were recovered and no single cluster dominated the simulation. The overall success of HMX-directed refinement (Fig. 5) suggests that de novo RNA structure refinement, based on easily obtained high-quality biochemical constraints, holds substantial promise for understanding structure-function interrelationships for RNA.
Experiments with the intact Tetrahymena group I intron illustrated how HMX analysis can characterize multiple stable structural intermediates within a single RNA population at single nucleotide resolution. HMX revealed that this RNA is stabilized by three sets of highly cooperative tertiary interactions. The first set corresponds to interactions between the P1–P2 helix and the rest of the folded RNA. The second corresponds to an extensive set of interactions that form between the P9-P7-P3-P8 domain and the P5-P4-P6 domain. The final set of interactions corresponds to intermolecular interactions that stabilize the P5-P4-P6 domain. This higher order domain structure was easily defined in a single set of experiments (Figs. 6 and 7) and is fully consistent with extensive prior characterization of this RNA.27,28
HMX is a simple, information-rich, and highly quantitative approach for analysis of the tertiary structure architecture of functionally important RNAs and provides a unique view of internal and closely packed RNA tertiary structure. Here, RNAs were partitioned based on size using gel electrophoresis; however, any strategy that separates functional from non-functional RNAs could be used, allowing HMX analysis to be implemented based on the ability of an RNA to interact with proteins or with other RNAs, or to perform catalysis and other functions.
Methods
RNA constructs
DNA templates for yeast tRNAAsp, the E. coli TPP and B. subtilis M-Box riboswitches, the Tetrahymena group I intron P546 domain, and the full length Tetrahymena group I intron template (425 nts) included 5' and 3' structure cassette flanking sequences13 and were generated by PCR. RNAs were transcribed in vitro [1 mL; 40 mM Tris (pH 8.0), 10 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine, 0.01% (v/v) Triton X-100, 4% (w/v) poly(ethylene) glycol 8000, 2 mM each NTP, 50 µL PCR-generated template, 0.1 mg/mL T7 RNA polymerase; 37 °C; 4 h] and purified by denaturing polyacrylamide gel electrophoresis [8% polyacrylamide, 7 M urea, 29:1 acrylamide:bisacrylamide, 0.4 mm × 28.5 cm × 23 cm gel; 32 W, 1.5 h]. RNAs were excised from the gel, recovered by passive elution overnight at 4 °C, and precipitated with ethanol. The purified RNAs were resuspended in 50 µL TE and stored at −20 °C.
RNA modification for molecular interference
RNA was mixed with its [32P]-labeled equivalent, denatured by heating to 90 °C for 2 min [32 µL; 30 pmol unlabeled RNA, 106 dpm 5'-[32P]-radiolabeled RNA, 100 mM HEPES (pH 8.0)], added to an NMIA solution (1.3 µL, 0.4 M in DMSO), and allowed to react at 95 °C for 5 min. This modification process was repeated three times and, after the third modification, the sample was placed on ice. For the full length Tetrahymena group I intron RNA, 60 pmol unlabeled RNA were used and the modification procedure was implemented twice. A no-modification control reaction was performed identically using neat DMSO. Any water evaporated during the modification was replaced to bring the volume to 36 µL (TPP samples were brought to 32 µL). For experiments in which band populations were quantified and visualized, 100,000 dpm of 5'-[32P]-radiolabeled RNA was used per condition; gels were visualized by phosphorimaging.
RNA folding, structural partitioning, and adduct detection
Modified RNA was treated with 4 µL 10× folding buffer (100 mM MgCl2, 1 M NaCl), and incubated at 37 °C for 30 min. Folding of the TPP riboswitch RNA was similar except that the RNA was incubated in folding buffer at 37 °C for 10 min, after which the TPP ligand (4 µL, 50 mM) was added and the sample was incubated at 37 °C for 20 min. The 40 µL of folded RNA sample was immediately added to equal volume of an 80% glycerol solution containing bromophenol blue and xylene cyanol and resolved on a non-denaturing polyacrylamide gel [8% polyacrylamide, 19:1 acrylamide:bisacrylamide, 0.5×TB (45 mM Tris, 45 mM boric acid), 50 mM NaCl, 5 mM Mg2Cl; 0.4 mm × 28.5 cm × 23 cm gel; 20 W, 8 h]. The gel was run in a cold room at 4 °C to ensure that the gel temperature did not increase above 37 °C. The anode and cathode buffer wells were periodically refreshed to maintain ion homostasis. Bands were visualized by exposing the gel to film (Kodak BioMax) for 1 hr. The film was used as a template to guide excision of the unfolded and folded band from the gel. Samples were recovered by passive elution overnight at 4 °C and were purified by ethanol precipitation and resuspended in 10 µL water. 2'-O-ester adducts in each band were detected by reverse transcription using fluorescently labeled primers, and resolved by capillary electrophoresis, as outlined previously.13,30
The HMX score
HMX experiments measure the functional partitioning of the folded, SF(i), and unfolded, SU(i), RNA ensembles in the presence of an 2'-O-ester adduct, where SF(i) and SU(i) are the intensity of the folded and unfolded bands for nucleotide i. The HMX score is:
where the coefficient α reflects the relative populations of RNAs with modifications at each nucleotide i (given that the absolute number of modifications in the ensemble cannot be determined). We estimated α by assuming that the total amount of adduct at each nucleotide position is roughly the same by minimizing the ratio between the average total modifications over the corresponding standard deviation, 〈SU (i) + αSF(i)〉/δ(SU (i) + αSF (i)) where the average and standard deviation are taken over all nucleotide positions:
Physical separation of folded and unfolded ensembles is not completely quantitative. The folded ensemble was generally well defined, but there was some folded-like RNA in the unfolded ensemble. We took this into account by subtracting the folded reactivity profile from the nonnative one:
The contribution of the folded state was most pronounced for nucleotides with low adduct reactivity in the unfolded ensemble; therefore, we estimated β by identifying regions with relatively low adduct reactivity in the unfolded ensemble that also had the highest correlation coefficients between the unfolded and folded ensembles (Supp. Fig. 2). The coefficient β was defined as the slope of the linear regression between unfolded and folded 2'-O-ester adduct intensity in these regions.
Modeling of adduct disruption of native RNA tertiary structure
The 2'-O-ester adducts were modeled as spheres (Fig. 4A). Hydrogen atoms were added using the Molprobity web service31 and the RNA model was extracted from a pdb file. Volume integrals were calculated using a Monte Carlo integration algorithm. The center of the adduct sphere was defined as a vector in the direction of the ribose C2'-O2' bond of length L from the ribose O2' position. Atoms from the originating and directly adjacent 5' and 3' nucleotides were excluded from the calculation. Clashes between atoms of the RNA and the center of the adduct sphere were assumed to be most disruptive. Thus, points for the Monte Carlo integration were sampled from a normal distribution with σ defined as the radius of the adduct; points are thus concentrated at the center of the adduct sphere. Points falling within the van der Waals radii of atoms in the PDB were scored as hits. Volume integrals converged after sampling 100,000 points at each nucleotide position.
HMX-directed structure refinement by DMD
DMD simulations comprised three steps. First, the RNA was folded from the linear sequence, constrained by the accepted canonical base pairing pattern. Second, we performed replica exchange DMD simulations with the additional tertiary structure constraints derived from HMX scores. Finally, we selected the 100 structures with the lowest energies and highest correlations between the structure and the experimentally-derived HMX scores. We incorporated HMX information in terms of solvent accessibility, using an approached previously developed to model hydroxyl radical probing.24 The relative solvent accessibility was interpreted as the number of contacts that sugar pseudoatoms have within predetermined distance from the center of each pseudoatom. Positions with high HMX scores were allowed a larger number of tertiary contacts than those with low scores.24 We assigned two biasing potentials based on the number of tertiary contacts as calculated from the HMX score. The first term is an attractive potential that collapses the RNA to achieve a compact structure. The second term is repulsion potential to prevent over-burial of a nucleotide when it exceeds the assigned number of threshold contacts. The minimum and maximum number of threshold contacts was calculated from a structure database to be 0.5 and 11, respectively.24 For any nucleotide exceeding the HMX scores below or above the threshold, we assigned values of 0.5 and 11.
Replica exchange DMD simulations and consensus structure modeling
We performed replica exchange DMD simulations for each RNA system using twelve replicas with temperatures of 0.200, 0.215, 0.230, 0.246, 0.262, 0.277, 0.293, 0.311, 0.330, 0.350, 0.375, and 0.400 kcal/(mol˙kB). We set each exchange event to occur at 1000 DMD time steps according to a Metropolis based Monte Carlo algorithm. For each replica, we performed a 5 × 105 DMD time step simulation. We then generated snapshots every 100 DMD time units; from these snapshots, we selected 1000 snapshots with the lowest energies, then calculated correlations between the number of tertiary contacts and the relative solvent accessibilities derived from the HMX scores and selected the 100 structures with lowest (negative) correlations. We then performed the same selection procedure in reverse order, first selecting the 1000 structures with the lowest correlations based on the experimentally-determined HMX scores and then selecting the 100 structures with lowest energies. These 200 structures were then ranked by energy and structure reactivity correlation coefficient and 100 structures were used to represent the final structural ensemble.24 These final ensembles were clustered by hierarchical clustering. For each RNA, the clustering cutoff was three quarters of the average RMSD as a function of RNA length; that is, the clustering cutoff was smaller for smaller RNAs with the maximum for larger RNAs equal to 4 Å.26
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (MCB-1121024 to K.M.W.).
References
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 4.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway L, Wickens M. Methods Enzymol. Academic Press. Inc; 1989. Modification interference analysis of reactions using RNA substrates; pp. 369–379. [DOI] [PubMed] [Google Scholar]
- 6.Clarke PA. RNA footprinting and modification interference analysis. In: Haynes S, editor. Methods Mol. Biol. Humana Press; 1999. pp. 73–91. [DOI] [PubMed] [Google Scholar]
- 7.Merryman C, Noller HF. RNA:protein interactions, a practical approach. Oxford University Press; Footprinting and modification-interference analysis of binding sites on RNA, in; pp. 237–253. [Google Scholar]
- 8.Ryder SP, Strobel SA. Nucleotide Analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 9.Strobel SA. A chemogenetic approach to RNA function/structure analysis. Curr. Opin. Struct. Biol. 1999;9:346–352. doi: 10.1016/S0959-440X(99)80046-3. [DOI] [PubMed] [Google Scholar]
- 10.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Pastor N, Weinstein H, Jamison E, Brenowitz M. A detailed interpretation of OH radical footprints in a TBP-DNA complex reveals the role of dynamics in the mechanism of sequence-specific binding. J. Mol. Biol. 2000;304:55–68. doi: 10.1006/jmbi.2000.4173. [DOI] [PubMed] [Google Scholar]
- 12.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 14.McGinnis JL, Dunkle JA, Cate JHD, Weeks KM. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westhof E, Dumas P, Moras D. Restrained refinement of two crystalline forms of yeast aspartic acid and phenylalaine transfer RNA. Acta. Cryst. A. 1988;44:112–123. [PubMed] [Google Scholar]
- 16.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dann CE, III, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and Mechanism of a Metal-Sensing Regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 18.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;2733:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharyya A, Murchie AIH, Lilley DMJ. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990;343:484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- 20.Lesnik EA, Guinosso CJ, Kawasaki AM, Sasmor H, Zounes M, Cummins LL, Ecker DJ, Cook PD, Freier SM. Oligodeoxynucleotides Containing 2'-O-Modified Adenosine: Synthesis and Effects on Stability of DNA:RNA Duplexes. Biochemistry. 1993;32:7832–7838. doi: 10.1021/bi00081a031. [DOI] [PubMed] [Google Scholar]
- 21.Lesnik EA, Freier SM. What affects the effect of 2“-alkoxy modifications? 1. Stabilization effect of 2-”methoxy substitutions in uniformly modified DNA oligonucleotides. Biochemistry. 1998;37:6991–6997. doi: 10.1021/bi972995c. [DOI] [PubMed] [Google Scholar]
- 22.Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J. Am. Chem. Soc. 2009;131:2541–2546. doi: 10.1021/ja805460e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–1173. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding F, Lavender CA, Weeks KM, Dokholyan NV. Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat. Methods. 2012;9:603–608. doi: 10.1038/nmeth.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavender CA, Ding F, Dokholyan NV, Weeks KM. Robust and generic RNA modeling using inferred constraints: a structure for the Hepatitis C virus IRES pseudoknot domain. Biochemistry. 2010;49:4931–4933. doi: 10.1021/bi100142y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajdin CE, Ding F, Dokholyan NV, Weeks KM. On the significance of an RNA tertiary structure prediction. RNA. 2010;16:1340–1349. doi: 10.1261/rna.1837410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. How the Group I Intron Works: A Case Study of RNA Structure and Function. In: Chech TR, Gesteland RF, Atkins JF, editors. The RNA World. 3rd. New York: Cold Spring Harbor Laboratory Press; 2005. pp. 133–205. [Google Scholar]
- 29.Golden BL. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 30.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2012;19:63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acid Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lescoute A, Westhof E. The interaction networks of structured RNAs. Nucleic Acid Res. 2006;34:6587–6604. doi: 10.1093/nar/gkl963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.