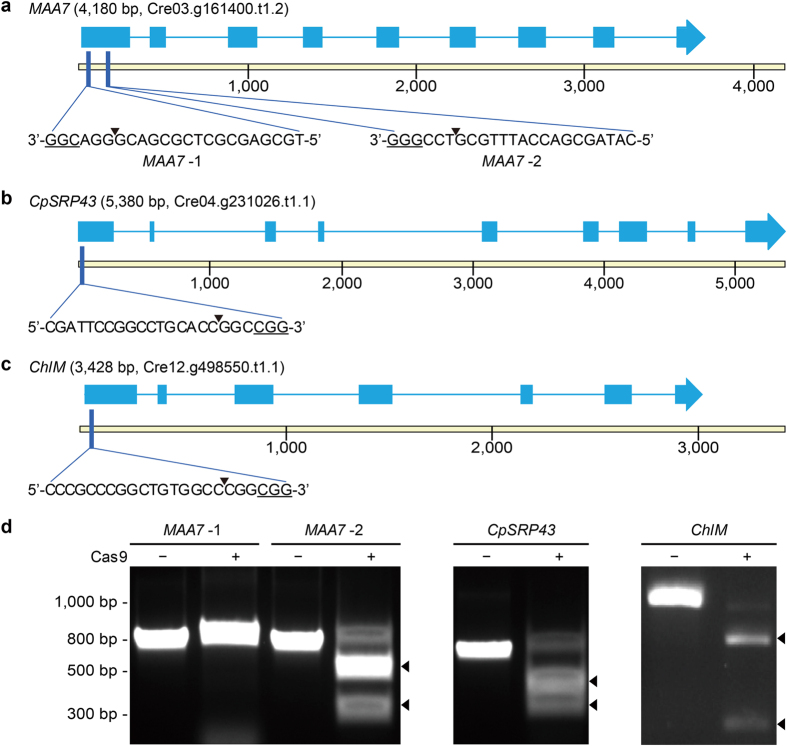
Figure 1. Map of CRISPR/Cas9 target sites in the MAA7, CpSRP43 and ChlM loci, and in vitro cleavage of target DNAs by sgRNAs and the Cas9 protein.
(a–c) Molecular maps of the MAA7 (a), CpSRP43 (b) and ChlM (c) loci with the CRISPR/Cas9 target sites indicated. CDS sequences are indicated as blue boxes, together with the sgRNA target sequences, the cut sites (arrowheads) and the associated PAM sites (5′-NGG-3′, underlined). Target sites of MAA7 were written in the 3′-to-5′ orientation, since the negative strand was targeted by sgRNAs. (d) The activities of the Cas9 protein and individual sgRNAs were tested in vitro. The Cas9-degraded products of the amplified fragments are indicated by arrowheads, with degraded products of 488 and 295 bp, 418 and 377 bp, and 736 and 274 bp identified for MAA7, CpSRP43 and ChlM, respectively.