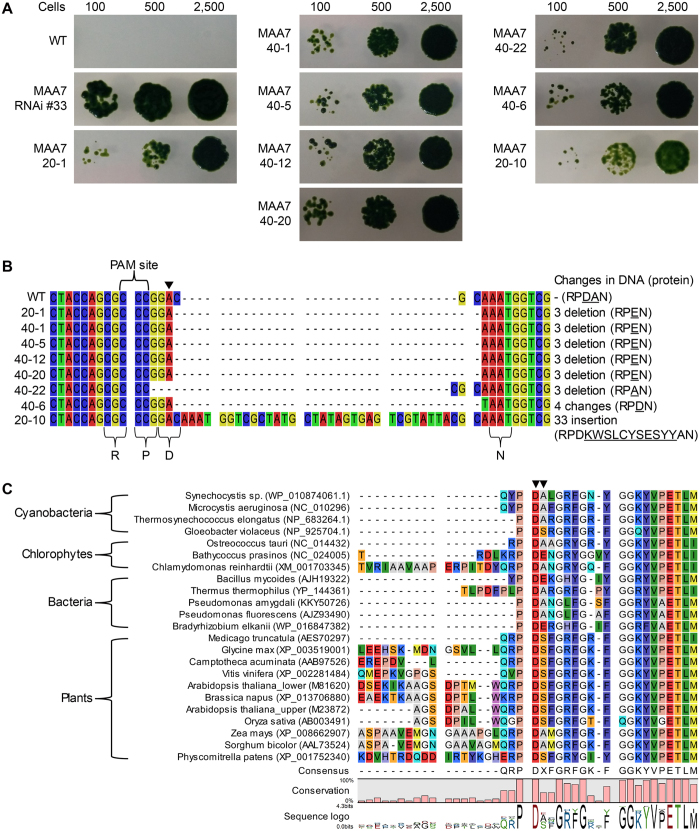
Figure 2. Auxotrophic selection of MAA7 mutants and their sequence analyses.
(A) The isolated maa7 mutants were grown in the presence of 5-FI with tryptophan supplementation, and cell growth was examined by spot-test loading of 100, 500, and 2500 cells. The RNAi mutant, MAA7 RNAi #33, was used as a reference strain. (B) Small indels are produced by CRISPR/Cas9 at the Cas9 cut site of the MAA7 locus. Sequencing of genomic DNA fragments encompassing the cut site revealed eight small indels at the cut site (at the 3′ side of the arrow head). The protein sequences of the target region (the RPDAN motif) are listed on the right side, and the altered sequences are underlined. (C) Alignments of the N-terminal side of the TSB sequences from various organisms representing three kingdoms. GenBank IDs are included to facilitate identification. Mutated sequences are marked with arrowheads.