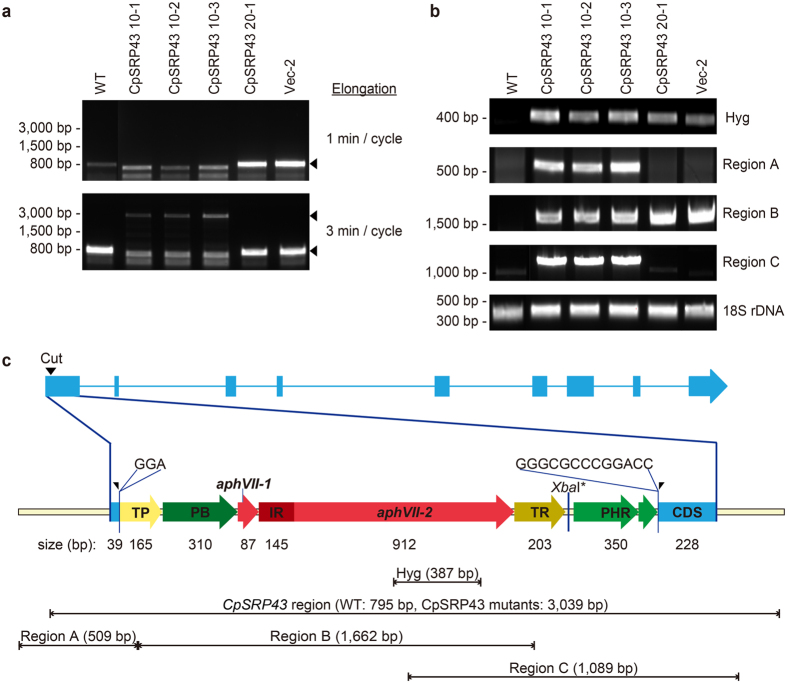
Figure 5. NHEJ-mediated knock-in of the co-transformed vector at the CRISPR/Cas9 cut site in the CpSRP43 locus.
(a) PCR amplification of the CpSRP43 regions was performed using different elongation times. Amplification of fragments encompassing the cut site failed under a short elongation cycle (upper panel), but improved when we used a longer cycle (lower panel). (b) Various vector sequences were confirmed by PCR. The amplicon sizes of the fragments encompassing the hygromycin resistance region, region A, region B, region C, and the 18S rDNA were 387 bp, 509 bp, 1,662 bp, 1,089 bp and 380 bp, respectively. The specific primers used in this study are listed in Table S1. (c) Molecular map of the CpSRP43 locus and the integrated vector sequences. An insertion of 3 bp (GGA) was identified next to the 5′ side of the Cas9 cut site (left-half arrowhead). Considerable vector sequences (2,650 bp) were inserted at the cut site, followed by an additional 13 bp of unknown origin (GGGCGCCCGGACC) at the 3′ side of the cleaved site (right-half arrowhead). All of the CpSRP43 mutants (10-1, 10-2 and 10-3) contained the same sequences at the Cas9 cut site in the CpSRP43 locus. Abbreviations: TP, PsaD terminator; PB, beta2-tub promoter; IR, RBCS2 intron; TR, RBCS2 terminator; and PHR, HSP70A-RBCS2 promoter.