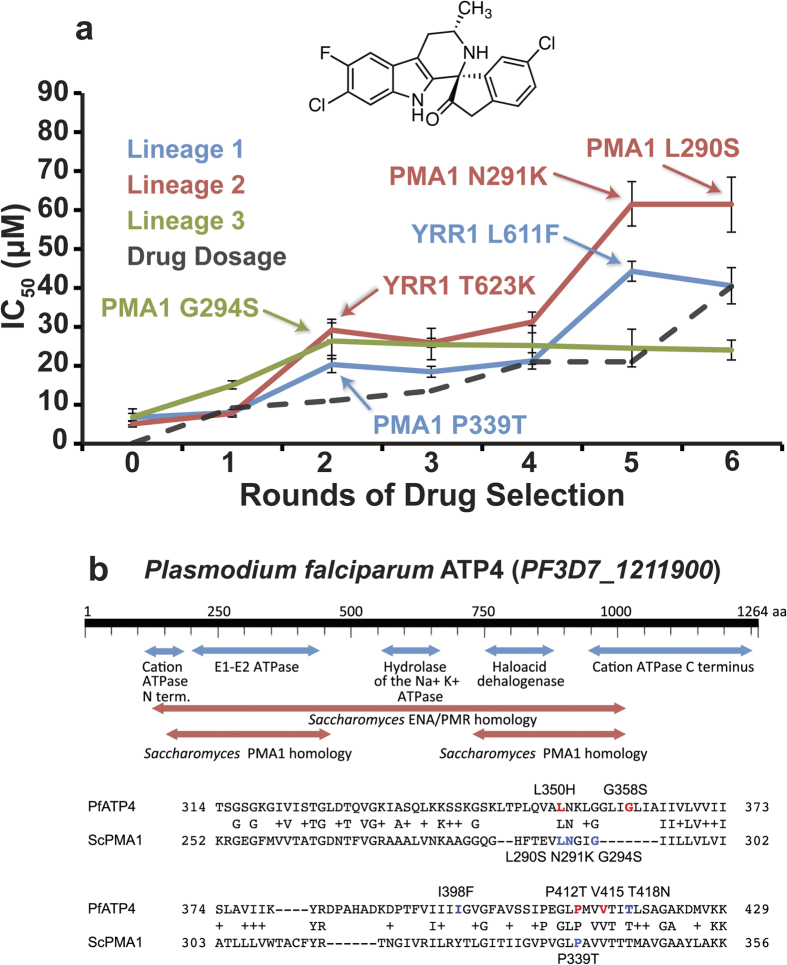
Figure 1. KAE609 directed evolution produces mutations in ScPMA1.
(a) An IC50 analysis of ABC16-Monster resistant lines showed that resistance developed over multiple rounds of drug selection. Sanger sequencing of ScPMA1 and ScYRR1 at each round was used to determine when each mutation (highlighted) arose in its respective lineage. (b) Alignment of PfAtp4p to ScPma1p (S288c) using PSI-BLAST (Position-Specific Iterated BLAST)68. The figure shows conserved domains (PfAtp4p amino acid 139–462:ScPMA1 87–389; PfATP4 744–1032:ScPMA1 527–784). ScPma1p and PfAtp4p are homologous only in the E1-E2 ATPase domain. The alignments of PfAtp4p amino acids 314–429 and ScPma1p 252–356 are shown below. Residues that confer resistance in Plasmodium when mutated are colored based on the compound class used: red for dihydroisoquinolones and blue for spiroindolones (see38 for a review).