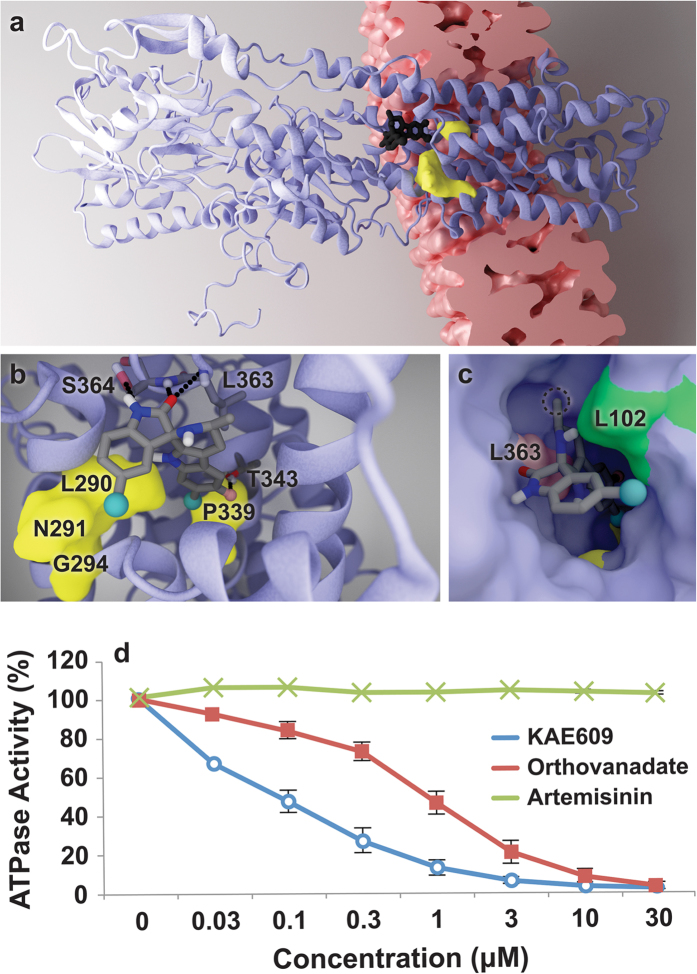
Figure 3. Functional and docking analyses support the hypothesis that KAE609 is a direct PfAtp4p inhibitor.
(a–c) Illustrations of the ScPma1p homology model and KAE609 docked pose. (a) The docked ligand is shown in black. Amino acids associated with evolved resistance are shown in yellow. The lipid-bilayer location was predicted using PDBID 1MHS69, CHARMM-GUI70, and the OPM database71. (b) The ligand with predicted hydrogen-bond partners (Ser364, Thr343, and Leu363). (c) The receptor in surface representation. Leu102 (green) is predicted to form hydrophobic contacts with KAE609. The position of Leu363 (pink) explains why the chirality at the 3′ carbon atom is critical to potency; if inverted, the attached methyl group (circled) would clash sterically with Leu363. (d) KAE609 inhibition of ScPMA1p in the vesicle-based assay25. Orthovanadate, a non-specific ATPase inhibitor, and artemisinin, an unrelated antimalarial, are included as controls. Two independent experiments were performed.