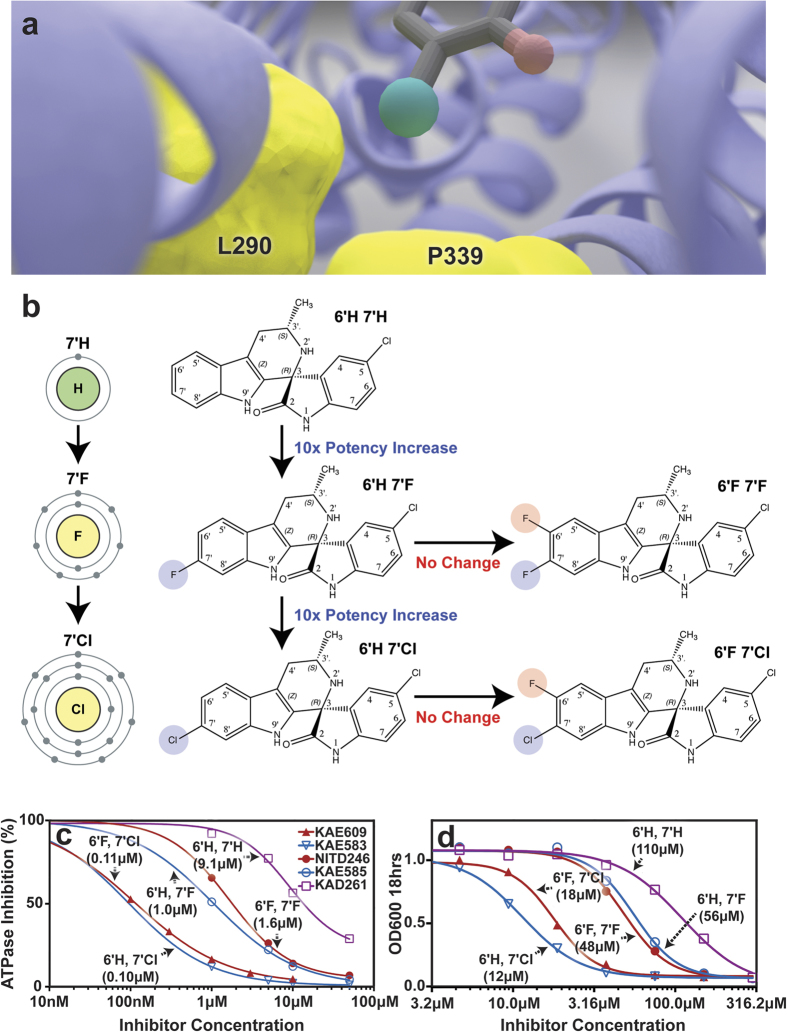
Figure 4. KAE609 halogenation.
(a) An illustration of the predicted binding pose, which positions the 7′ chlorine atom (green) between two residues that were altered during directed evolution (L290S and P339T). Unlike the polar mutant residues, the nonpolar wild-type residues may stabilize the interaction with the 7′ chlorine atom. (b) In yeast, halogenation at the 7′ position (blue) has a substantial impact on potency, but halogenation at the 6′ position (red) has little impact. Note that KAE609 is the molecule designated 6′F 7′Cl. (c) The blue, red, and purple curves indicate 7′, 6′/7′, and no halogenation. In the cell-free assay, 7′ chlorination yields ~10- and ~100-fold greater potency than fluorination and no halogenation, respectively. (d) Structure-activity relationships showing correlation between direct inhibition of ScPMA1p in the vesicle-based assay25 and activity in a whole-cell yeast replication assay (see methods). The in vitro IC50 values against blood-stage P. falciparum NF54 (in nM) were 13 ± 2.2 (6′H 7′H), 4.3 ± 0.4 (6′H 7′F), 0.33 ± 0.06 (6′F 7′F), 5.6 ± 0.7 (6′H 7′Cl), and 1.2 ± 0.2 (6′Cl 7′F, KAE60972. The following naming conventions were used by Yeung et al. in their paper concerning spiroindolone SAR: 6′H 7′H = (+)-1, 6′H 7′F = (+)-5, 6′F 7′F = (+)-6, 6′H 7′Cl = (+)-3, and 6′Cl 7′F (KAE609) = (+)-726.