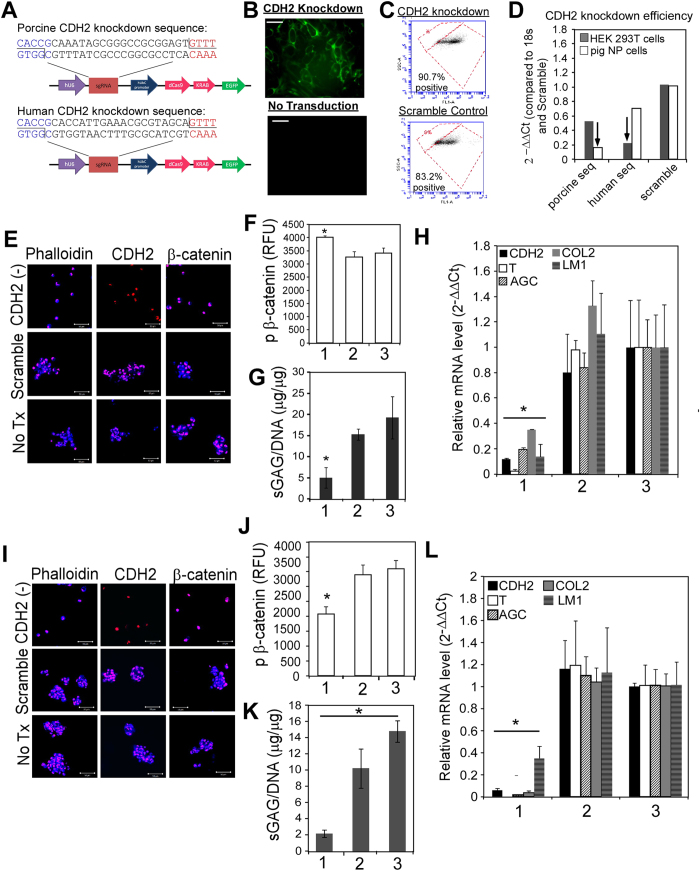
Figure 3. CDH2 knockdown (via CRISPRi) prevents cell cluster formation on soft, PEG-LM in porcine and human NP cells, with associated loss of juvenile NP phenotype.
(A) CRISPRi construct sequence. (B) Representative images of successful CDH2 knockdown analyzed by fluorescence microscopy. (C) Validation of successful CDH2 knockdown via flow cytometry. (D) Quantification of CDH2 knockdown via qRT-PCR; arrows indicate successful CDH2 knockdown using species-specific CRISPRs (porcine seq = porcine sequence, human seq = human sequence). (E) Representative immunostaining for phalloidin, CDH2 and β-catenin in porcine NP cells after CDH2 knockdown, compared to scramble control and no treatment (blue = protein, pink/red = cell nuclei, scale bar = 50 μm). (F) Quantification of total phosphorylated β-catenin levels in porcine NP cells (1 = CDH2 (−), 2 = scramble, 3 = no treatment). (G) Changes in sGAG in CDH2 knockdown porcine NP cells on soft. (H) Changes in gene expression for CDH2 knockdown porcine NP cells on soft, compared to stiff (CDH2 = N-cadherin, T = brachyury, LM1 = laminin, COL2 = type II collagen, AGC = aggrecan; * denotes that all genes are significantly different from scramble and no treatment controls) (I–L) same as (E–H) respectively, but with CDH2 knockdon in juvenile human NP cells (For all studies: 2-way ANOVA with Tukey’s post-hoc analysis, *p < 0.05).