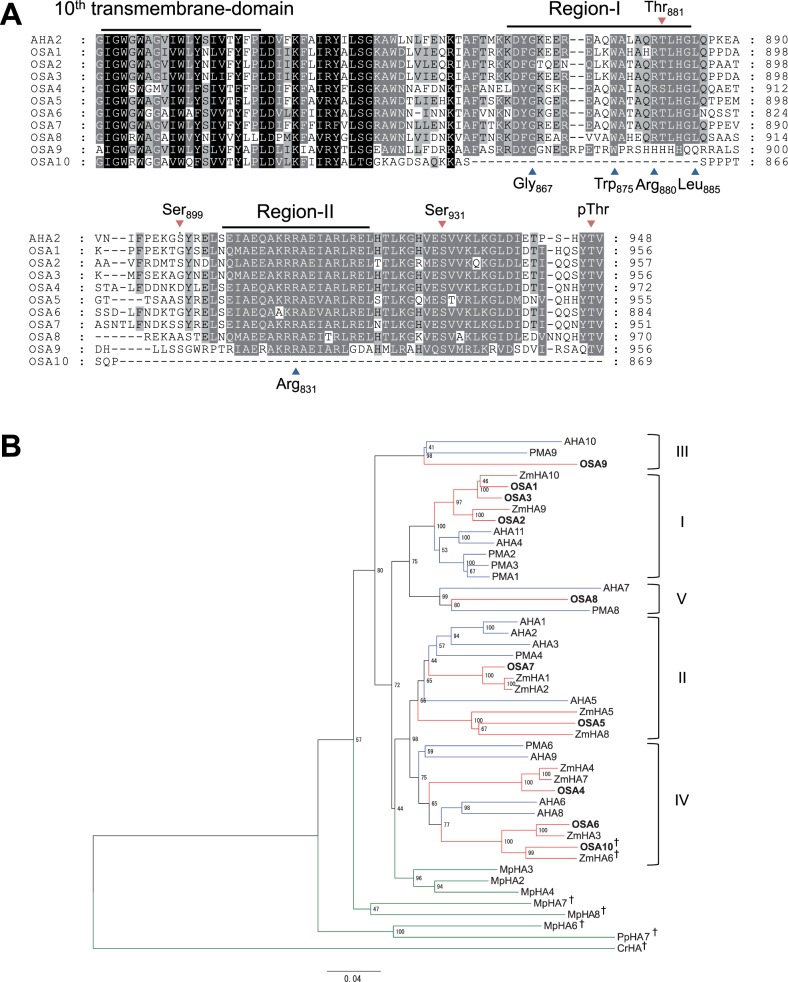
Fig. 2.
Sequential and phylogenetic analysis of H+-ATPases in rice. (A) Sequence alignment of the C-terminal inhibitory domain of Oryza sativa H+-ATPases (OSAs) and Arabidopsis AHA2. The 10th transmembrane domain and the inhibitory motif (Region-I and Region-II) within the C-terminal inhibitory domain are shown. Blue arrowheads below the sequence indicate amino acids that are critical for the function of the inhibitory domain of AHA2 (Axelsen et al. 1999). Red arrowheads over the sequence indicate phosphorylation target sites in AHA2 (Fugisang et al. 2007, Niittylä et al. 2007, Haruta et al. 2014). (B) Phylogenetic tree of H+-ATPases of rice, maize (ZmHAs), Arabidopsis (AHAs), tobacco (PMAs), M. polymorpha (MpHAs), P. patens (PpHA) and C. reinhardtii (CrHA). The phylogenetic tree was constructed using the full-length amino acid sequences of H+-ATPases. The scale bar indicates 0.04 amino acid substitutions per site. Blue, red and green nodes represent H+-ATPases of dicots, monocots and others, respectively. CrHA was used as an outgroup. Daggers indicate non-pT-type H+-ATPases. Roman numerals indicate subfamilies defined by Arango et al. (2003). PMA5, PMA7, MpHA1 and MpHA5 were not incorporated into this analysis because full-length sequences were not available.