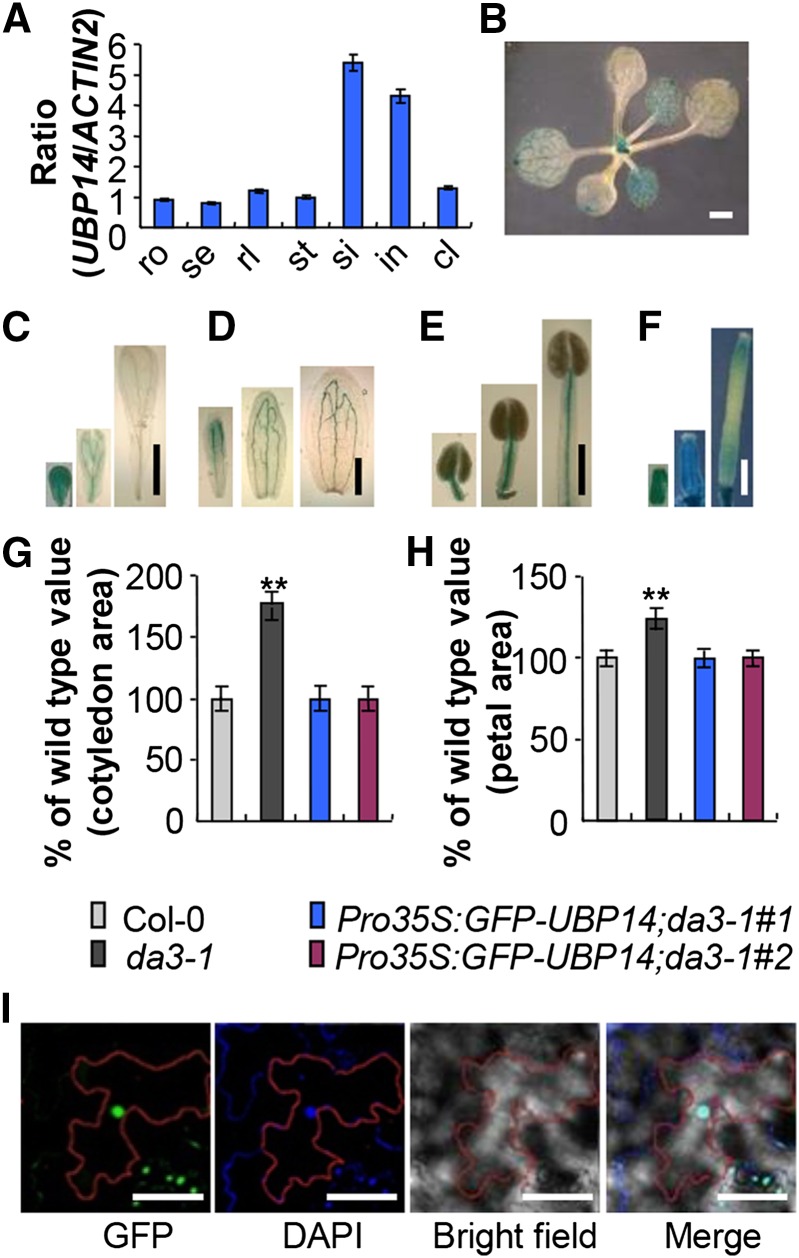
Figure 3.
Expression Pattern and Subcellular Localization of UBP14.
(A) Quantitative real-time RT-PCR analysis of DA3 expression in the indicated Arabidopsis tissues. Total RNA was isolated from roots (ro), seedlings (se), rosette leaves (rl), stems (st), siliques (si), inflorescences (in), and cauline leaves (cl). Data shown are means ± sd of three biological replicates.
(B) to (F) UBP14 expression activity was monitored by ProUBP14:GUS transgene expression. Twenty GUS-expressing lines were observed, and all showed a similar pattern, although they differed slightly in the intensity of the staining. Histochemical analysis of GUS activity in 12-d-old seedlings (B), the developing petals (C), the developing sepals (D), the developing stamens (E), and the developing carpels (F).
(G) Cotyledon area of seedlings of Col-0, da3-1, Pro35S:GFP-UBP14;da3-1#1, and Pro35S:GFP-UBP14;da3-1#2 (n = 55). Pro35S:GFP-UBP14;da3-1 represents da3-1 transformed with Pro35S:GFP-UBP14.
(H) Petal area of Col-0, da3-1, Pro35S:GFP-UBP14;da3-1#1, and Pro35S:GFP-UBP14;da3-1#2 (n = 65). Pro35S:GFP-UBP14;da3-1 is da3-1 transformed with Pro35S:GFP-UBP14.
(I) Subcellular localization of GFP-UBP14 in epidermal cells of Pro35S:GFP-UBP14;da3-1 leaves. GFP fluorescence of GFP-UBP14, DAPI staining, and bright-field and merged images (from left to right) are shown. The red lines indicate the outline of an epidermal cell.
Values in (G) and (H) are given as means ± sd relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test).
Bars = 1 mm in (B), 1 mm in (C) to (F), and 50 μm in (I).