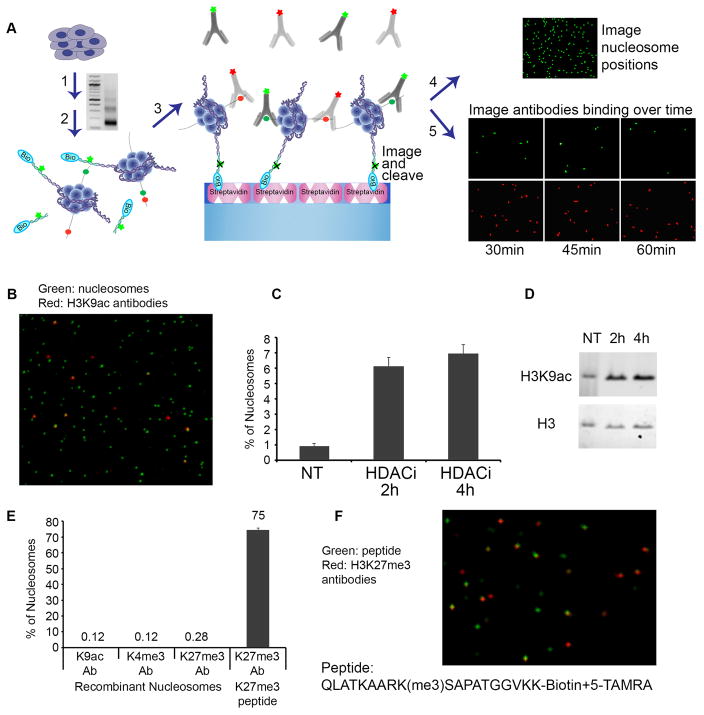
Fig. 1. Single-molecule detection of post-translational modifications on nucleosomes.
(A) Experimental scheme: (1) Nucleosomes from cells are prepared by Micrococcal Nuclease (MNase) digestion. Gel depicts nucleosomal DNA fragments of expected lengths; (2) Free DNA ends are ligated to fluorescent, biotinylated oligonucleotide adaptors; (3) Adaptor-ligated mono-nucleosomes are purified on a glycerol gradient and captured on PEG-streptavidin coated slides. (4) Nucleosome positions on the surface are imaged by TIRF microscopy, and then the fluorophore is cleaved from the adaptor. (5) Attached nucleosomes are incubated with fluorescently-labeled antibodies to histone modifications. Time-lapse images detect repeated binding and dissociation events and are integrated to score modified nucleosomes. (B-D) HEK293 cells were treated with HDAC inhibitor. (B) Single-Molecule detection of labeled nucleosomes (Alexa555, green) bound by labeled H3K9ac antibodies (Alexa647, red). (C) Proportion of nucleosomes marked by H3K9ac under each condition is determined by single-molecule counting. (D) Western blot confirms increased H3K9ac in treated cells. (E–F) Recombinant unmodified nucleosomes and H3K27me3-modified peptide were probed with the indicated antibodies. (F) Single-molecule detection of labeled H3K27me3 peptide (TAMRA, green) with labeled H3K27me3 antibodies (Alexa647, red) at a single time point.