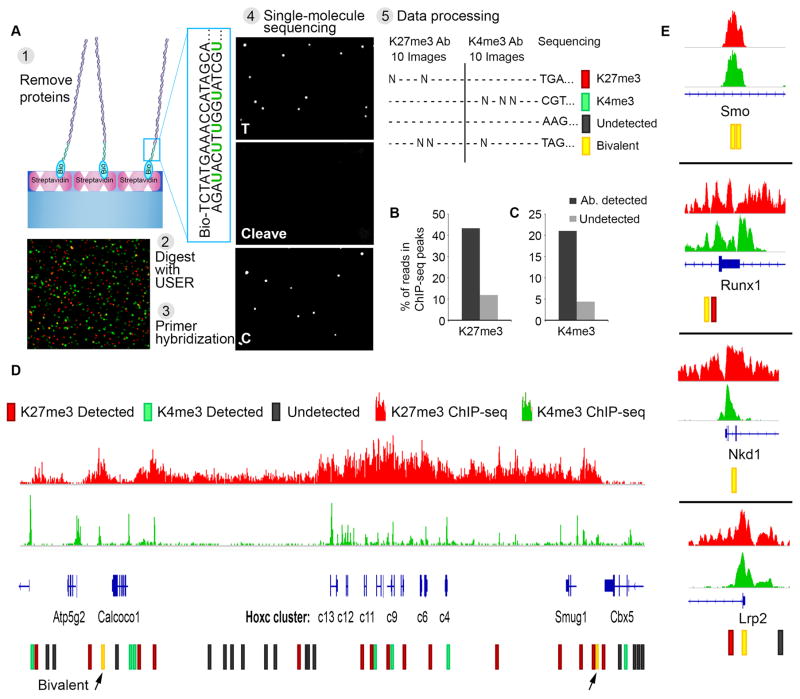
Fig. 4. Single-molecule sequencing determines genomic positions of modified nucleosomes.
(A) Experimental scheme: (1) Nucleosomes are captured and probed for their modification state, as in Fig. 1A. Histones are evicted by increasing salt concentration. (2) The enzyme USER is applied to excise uracil bases incorporated into the non-biotinylated adaptor strand and expose a known sequence (3) Complementary primer is hybridized to the adaptor. Image shows single molecule detection of nucleosomal DNA (Alexa647, red) and primer (Alexa555, green). (4) Direct single-molecule DNA sequencing-by-synthesis (22). Images reflect two sequencing cycles: incorporation of thymine, cleavage of fluorophore and terminator, and incorporation of cytosine. (5) Data processing: for each x-y coordinate on the surface, sequence data is analyzed and integrated with the initial images scoring antibody binding and modification states of the corresponding nucleosomes. (B) Single-molecule reads were aligned to the genome. Plot indicates proportions of H3K27me3-modified nucleosome reads (‘detected’) or un-modified nucleosome reads (‘undetected’) that aligned to H3K27me3-enriched regions per conventional ChIP-seq. (C) Proportions of H3K4me3-modified nucleosome reads that aligned to H3K4me3-enriched regions per conventional ChIP-seq. (D) The HOXC gene cluster is shown along with H3K27me3 and H3K4me3 ChIP-seq tracks. Single-molecule reads that aligned to these regions are indicated, along with the modification status of the corresponding nucleosome. (E) Analogous data shown for other developmental loci for which bivalent nucleosomes were definitively identified.