Figure 1.
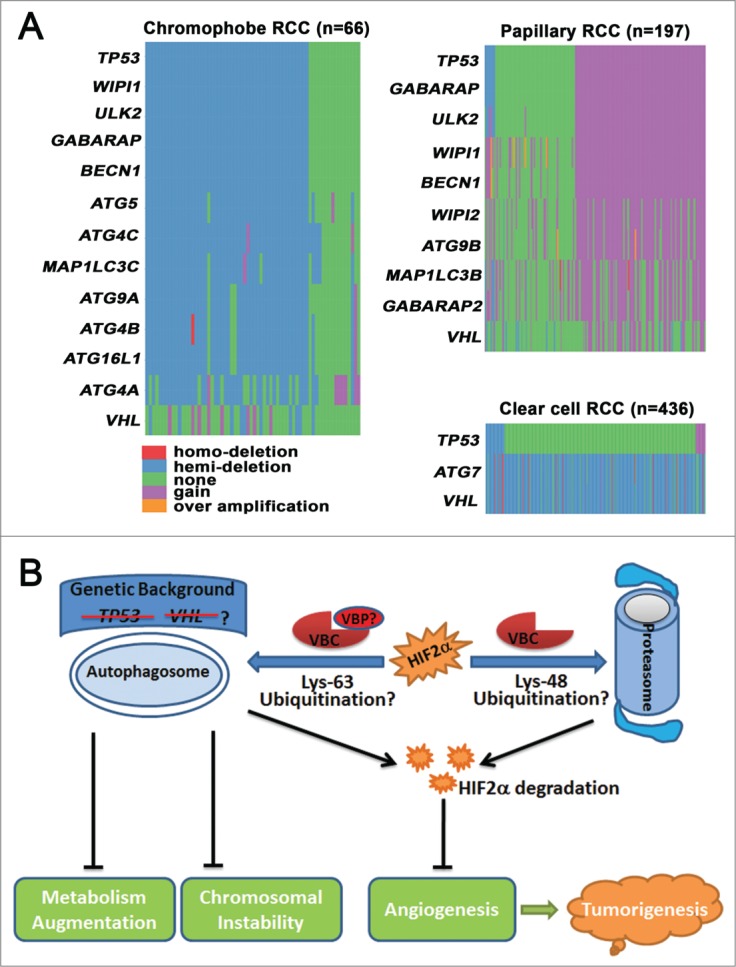
The function and genetic dysregulation of autophagy in renal cell carcinoma (RCC). (A) Copy number changes of autophagy-related genes (ATGs) in different subtypes of RCC. Copy number data for chromophobe RCC, papillary RCC, and clear cell RCC were obtained from The Cancer Genome Atlas (TCGA), and data processing was conducted with statistical computing tools. ATGs that were abnormal in more than 50% of cases are listed. The distribution of genes was drawn in the order of tumor protein P53 (TP53). (B) Autophagy suppresses tumorigenesis by degrading endothelial PAS domain-containing protein 1 (EPAS1, also known as HIF2α), stabilizing chromosomes, and moderating metabolism. Autophagy cooperates with the proteasome to degrade HIF2α and prevent angiogenesis. Lys48-linked polyubiquitination directs HIF2α to the proteasome for degradation, whereas Lys63-polyubiqiutinated HIF2α is subjected to autophagic degradation. VHL binding proteins (VBPs), such as VBP1 and sequestosome 1 (SQSTM1/p62), may preferentially promote Lys63-linked polyubiquitination and recruit Lys63-linked polyubiquitinated HIF2α to the autophagosome for degradation. Autophagy also suppresses renal tumorigenesis by preventing chromosomal instability and by moderating metabolism. The anticancer roles of autophagy may be more important in the absence of tumor suppressor genes, such as Von Hippel-Lindau (VHL) or TP53.
