Figure 1.
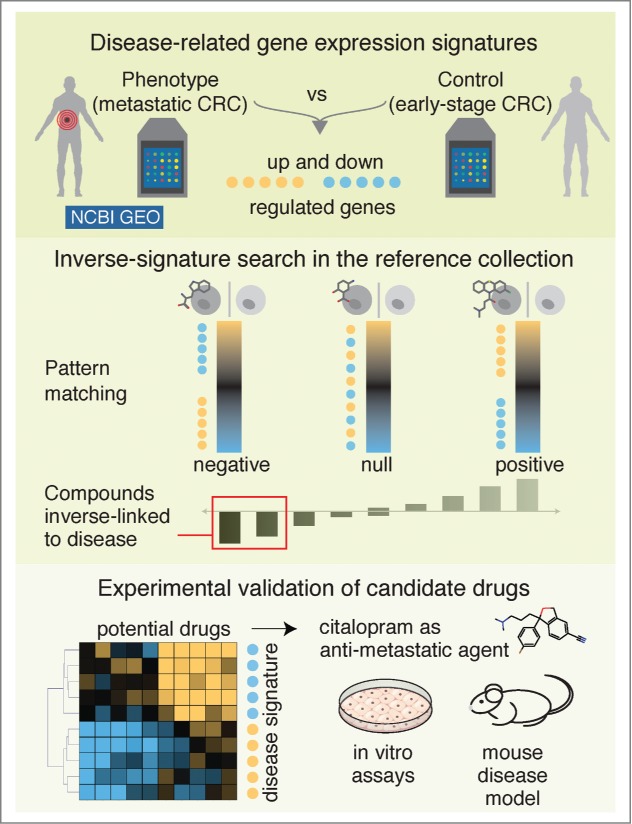
Workflow of the inverse-signature approach to identify potential therapeutic compounds. A disease transcriptional signature was extracted by comparing phenotype versus control (e.g., metastatic vs. primary colorectal cancer) gene expression profiles publicly available from Gene expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) at NCBI (National Center for Biotechnology Information). Next, the disease-related signature was queried against the reference collection of drug-induced gene expression profiles. Drug candidates with anti-correlated profiles were further refined using hierarchical clustering and the efficacy of selected candidates (e.g., citalopram) was experimentally tested using in vitro and in vivo disease models (e.g., colorectal cancer [CRC]).
