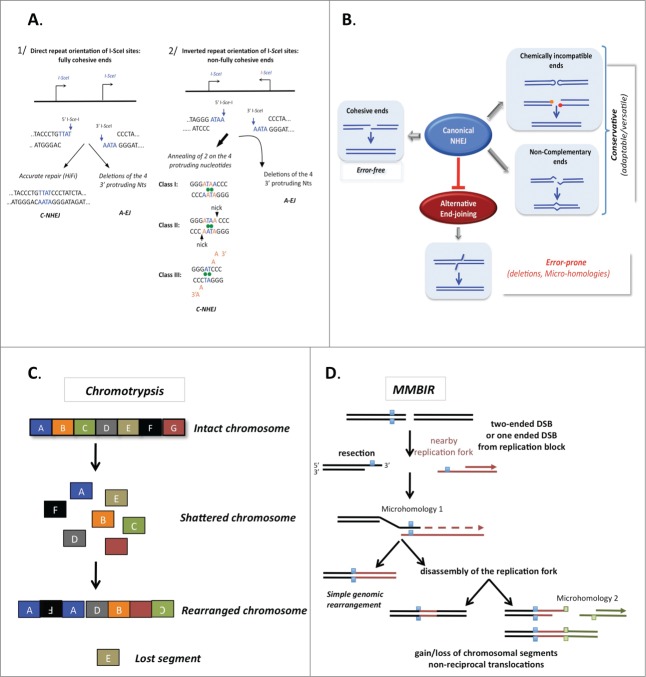
Figure 4.
The accuracy of end joining. (A) The fidelity of C-NHEJ on fully (left panel) or incompletely (right panel) complementary ends. Here, examples are given with ends generated by the meganuclease I-SceI. The cleavage sites are not palindromic therefore 2 I-SceI cleavage sites in the same orientations yield fully complementary ends (left panel), whereas 2 I-SceI cleavage sites in inverted orientation yield incompletely complementary overhangs (right panel). A-EJ leads to deletions with both types of DNA ends. C-NHEJ is mainly error-free on fully complementary ends and uses 3 classes of imperfect annealing (3 out of the 4 3′ protruding nucleotides generated by I-SceI cleavage) with non-fully complementary ends. Thus, C-NHEJ is conservative but adaptable for incompletely complementary ends. (B) The actual accuracy of end joining. A-EJ is highly mutagenic in all situations but it is blocked by C-NHEJ, which can act on imperfectly complementary ends. In situations producing non-ligatable ends, such as hairpins in V(D)J recombination or IR-induced multiple damages at DSBs, a preliminary processing step is required prior to end joining. Note that in these cases, diversity or mutagenesis is generated by the processing step rather than the end-joining machinery. (C) Chromothripsis. The religation of shattered chromosomes in small pieces (colored squares) leads to a combination of rearranged chromosomes in addition to amplification and loss of the small pieces. (D) A model for chromothripsis arising through MMBIR (microhomology-mediated break induced replication). Resection of the DNA double-strand end generates a 3′ overhang, which can anneal another DNA molecule (in red) through the use of microhomologies (blue squares), thus priming DNA synthesis (dotted arrow). This mechanism can lead to more complex rearrangements if it is coupled to multiple cycles of disassembly and template switches of the replication forks (right panel), still using microhomologies.