Abstract
Oncogene-associated metabolic signatures in prostate cancer, identified by an integrative analysis of cultured cells and murine and human tumors, suggest that AKT activation results in a glycolytic phenotype whereas MYC induces aberrant lipid metabolism. Heterogeneity in human tumors makes this simplistic interpretation obtained from experimental models more challenging. Metabolic reprogramming as a function of distinct molecular aberrations has major diagnostic and therapeutic implications.
Keywords: AKT signaling, cancer metabolism, mass spectrometry, MYC, prostate cancer
Emerging technologies offer opportunities to integrate high-throughput information from the genome, the transcriptome, and the proteome. More recently, metabolomics has also been expanding in scope, from its primary use in toxicology and microbiology to a new diagnostic role in cancer and other diseases.1,2
The concept that tumor cells undergo metabolic reprogramming is not novel. Warburg first discovered that cancer cells preferentially use aerobic glycolysis, reducing pyruvate into lactate in the presence of oxygen.3 However, human tumors are metabolically heterogeneous. Driver oncogenes in cancer cells use both oxidative metabolism and aerobic glycolysis for cell survival and growth. AKT and MYC, arguably the most prevalent driving oncogenes in prostate cancer, have been linked to glycolytic metabolism in previous studies. Interestingly, a differential metabolic fingerprint was identified in this Priority Report published in Cancer Research,4 associating activation of AKT1 with accumulation of aerobic glycolysis metabolites and overexpression of MYC with dysregulated lipid metabolism (Fig. 1).
Figure 1.
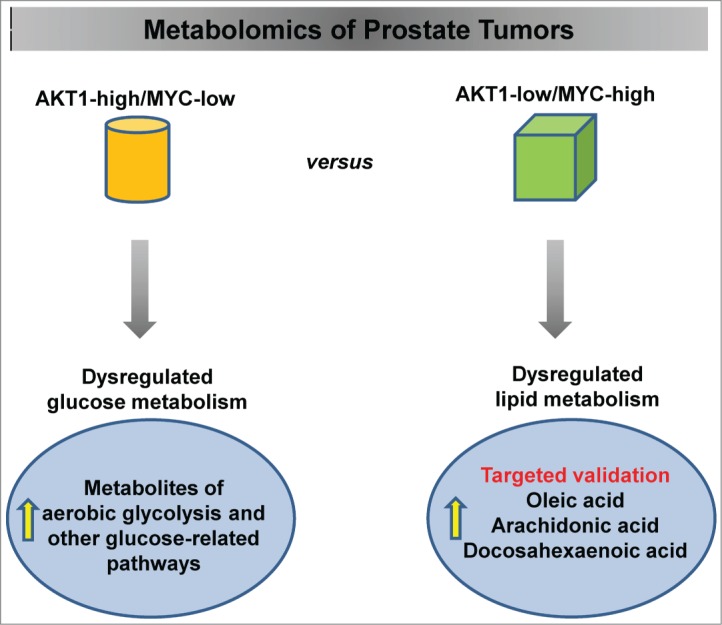
Identification of oncogene-specific signatures in prostate cancer by metabolomics. Activation of AKT1 signaling in prostate tumors results in the accumulation of intermediates and end products of glucose metabolism, whereas high MYC expression is associated with significantly increased levels of free fatty acids and glycerophospholipid metabolism intermediates. Selected lipids (oleic, arachidonic, and docosahexaenoic acids) were validated by quantitative mass spectrometry assays.
Prostate cancer does not always show the classic “glycolytic switch” seen in the majority of other solid tumors, and therefore is not always detectable with 18-fluorodeoxyglucose positron emission tomography (FDG-PET). Instead, an aberrant increase in de novo lipogenesis from glucose and glutamine is observed in the early stages of the disease, and is associated with tumor progression and shorter survival.5
MYC is frequently amplified in the late stages of prostate cancer, but is also overexpressed in the absence of a genetic lesion.6 Priolo et al. showed that MYC-driven cell lines and murine prostates are associated with deregulated lipid metabolism. However, the human data in this study showed metabolic heterogeneity in addition to genetic and signaling pathway heterogeneity. For example, the “pre-existing” metabolic state of normal prostate tissue seems to modulate metabolic reprogramming in tumors. Interestingly, whereas AKT1 activation in nontransformed prostate epithelial cells (human and mouse) induces an aerobic glycolytic signature, MYC-high tumors have a negative enrichment of glycolysis compared to adjacent benign prostate tissue. Lactate is not increased in phosphoAKT1-high tumors relative to normal prostate, yet it is significantly elevated compared to MYC-high tumors. In addition, high MYC expression in a phosphoAKT1-low context in human tumors is associated with decreased mRNA expression of the glucose transporter-1 (GLUT-1).4 MYC is known to induce aerobic glycolysis in certain preclinical models. However, although in our study metabolites of glycolysis were induced by acute overexpression of MYC in vitro, this aberration was not confirmed in transgenic mouse prostate (Lo-MYC) or MYC-high prostate tumors. It is tempting to speculate that MYC-driven tumors may rely more heavily on lipid metabolism. These findings suggest that the metabolic state in normal prostate tissue may play a role in modulating metabolic reprogramming by different oncogenes in transformed tissue.
Interestingly, both AKT1 and MYC induce expression of fatty acid synthase (FASN), which also plays an oncogenic role in prostate cancer,7 and levels of this enzymes are increased in nearly all prostate tumors relative to normal tissue. FASN could represent a common denominator of the 2 metabolic signatures, requiring a higher flux of glucose and/or glutamine to ultimately generate acetyl CoA, which is necessary for de novo lipogenesis.
Validation of untargeted metabolomics by quantitative assays for selected metabolites4 provides a valuable proof of concept for future large studies aimed at identifying biomarkers in human samples. In addition, the assessment of metabolite profiles in human sera, particularly through pathway analysis, could allow investigators to infer the genetic alterations driving prostate cancer, especially in the metastatic setting. Finally, metabolic profiling can guide the use of radiochemistry for in vivo imaging (e.g., 11C-acetate in lipogenic MYC-driven tumors).
Of note, metabolite levels do not necessarily reflect changes in expression levels of the enzymes that use them or generate them. Therefore, because the regulation of metabolism is complex, gene or protein expression of metabolic enzymes should be considered in conjunction with metabolic profiling in order to more accurately define metabolic aberrations in heterogeneous human tumors.
Conclusions and Perspectives
This study has several potential translational applications. Alterations of certain metabolic pathways in prostate tumors could be used diagnostically to infer the molecular phenotype of the tumor, or to monitor patients whose tumors have an established molecular profile. This is particularly important in a disease such as prostate cancer, in which metastatic disease in bone is not routinely biopsied. Furthermore, these results could lead to the discovery of radiopharmaceuticals to image prostate cancer patients noninvasively. Finally, enzymes in these metabolic pathways could represent therapeutic targets for prostate tumors that are addicted to the putative driving oncogenes. Thus, metabolic reprogramming as a function of distinct molecular aberrations can have major diagnostic and therapeutic implications.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by NIH/NCI grant 2R01CA131945, the DF/HCC SPORE in Prostate Cancer (NIH/NCI P50 CA90381), the Prostate Cancer Foundation, the DOD synergist idea development award 11498838 to M.L.
References
- 1.Chen R, Snyder M. Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med 2013; 5(1):73-82; PMID:23184638; http://dx.doi.org/ 10.1002/wsbm.1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012; 483(7390):479-83; PMID:22343889; http://dx.doi.org/ 10.1038/nature10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science 1956; 123(3191):309-14; PMID:13298683; http://dx.doi.org/ 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 4.Priolo C, Pyne S, Rose J, Regan ER, Zadra G, Photopoulos C, Cacciatore S, Schultz D, Scaglia N, McDunn J., et al.. AKT1 and MYC induce distinctive metabolic fingerprints in human prostate cancer. Cancer Res 2014; PMID:25322691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zadra G, Photopoulos C, Loda M. The fat side of prostate cancer. Biochim Biophys Acta 2013; 1831(10):1518-32; PMID:23562839; http://dx.doi.org/ 10.1016/j.bbalip.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh CM, Bieberich CJ, Dang CV, Nelson WG, Yegnasubramanian S, De Marzo AM. MYC and prostate cancer. Genes Cancer 2010; 1(6):617-28; PMID:21779461; http://dx.doi.org/ 10.1177/1947601910379132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Migita T, Ruiz S, Fornari A, Fiorentino M, Priolo C, Zadra G, Inazuka F, Grisanzio C, Palescandolo E, Shin E., et al., Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst 2009; 101(7):519-32; PMID:19318631; http://dx.doi.org/ 10.1093/jnci/djp030 [DOI] [PMC free article] [PubMed] [Google Scholar]


