Abstract
In plants, vacuolar H+-ATPase (V-ATPase) activity acidifies both the trans-Golgi network/early endosome (TGN/EE) and the vacuole. This dual V-ATPase function has impeded our understanding in how the pH homeostasis within the plant TGN/EE controls exo- and endocytosis. Here, we show that the weak V-ATPase mutant deetiolated3 (det3) displayed a pH increase in the TGN/EE, but not in the vacuole, strongly impairing secretion and recycling of the brassinosteroid receptor and the cellulose synthase complexes to the plasma membrane, in contrast to mutants lacking tonoplast-localized V-ATPase activity only. The brassinosteroid insensitivity and the cellulose deficiency defects in det3 were tightly correlated with reduced Golgi and TGN/EE motility. Thus, our results provide strong evidence that acidification of the TGN/EE, but not of the vacuole, is indispensable for functional secretion and recycling in plants.
Plant exo- and endocytic pathways converge at the trans-Golgi network/early endosome (TGN/EE) compartment where different cargos are sorted to further destinations1,2. In animal and yeast cells, acidification of intracellular organelles is crucial for the function of the secretory and endocytic pathways and requires proton pumping activity of the vacuolar H+-ATPases (V-ATPase)3–5. The V-ATPase is conserved across species and consists of multiple subunits that are organized in a cytosolic V1 domain, which is important for the ATP hydrolysis (including A, B, C, D, E, F, G, and H subunits), and an integral membrane V0 domain, which forms the proton pore (including a, d, c, c" and e subunits)3. In Arabidopsis thaliana, the V-ATPase activity is associated with both the TGN/EEs and the tonoplast that are marked by the differential localization of the membraneVHA-a1, VHA-a2 and VHA-a3 isoforms1,6,7. The vha-a3 mutant and the vha-a2 vha-a3 double mutant that lack the tonoplast V-ATPase activity do not display severe defects in cell expansion, whereas the inducible inhibition of the TGN/EE-localized VHA-a1 isoform constrains it7,8. Treatment with the V-ATPase inhibitor concanamycinA (ConcA) resulted in loss of the TGN/EE identity and interfered with the trafficking of endocytic and secretory cargos1,2. Given the differential localization of the V-ATPases, the reduced cell expansion has been concluded to be caused by defects in TGN/EE compartments rather than in the vacuole8, but the nature of these defects has not been clarified. In contrast, the cytosolic V-ATPase subunit C (VHA-C), encoded by the single-copy VHA-C/DEETIOLATED3 (DET3) gene, is required for V-ATPase activity at the TGN/EEs and at the vacuole9. A knockdown allele of DET3 displayed pleiotropic phenotypes including impaired cell expansion, photomorphogenesis in the dark, ectopic lignification, reduced sensitivity to brassinosteroids (BRs) and reduced cellulose levels8–11. A clear explanation for those phenotypes is currently lacking. BRs are sensed at the plasma membrane by the BRASSINOSTEROID-INSENSITIVE1 (BRI1) receptor that transits through the TGN/EE during secretion and endocytosis en route to the vacuole and also undergoes recycling1,2,12,13. Cellulose is synthesized at the plasma membrane by cellulose synthase A (CesA) complexes that are assembled in the Golgi, or the endoplasmic reticulum, and delivered to the plasma membrane where they become activated14–17. Our results reveal that the det3 mutant has a reduced ability to acidify the TGN/EE, but not the Golgi and the vacuole, leading to secretion and recycling defects that contribute to BR insensitivity and cellulose deficiency.
Results
The increased pH in TGN/EE of det3 caused low Golgi and TGN/EE motility
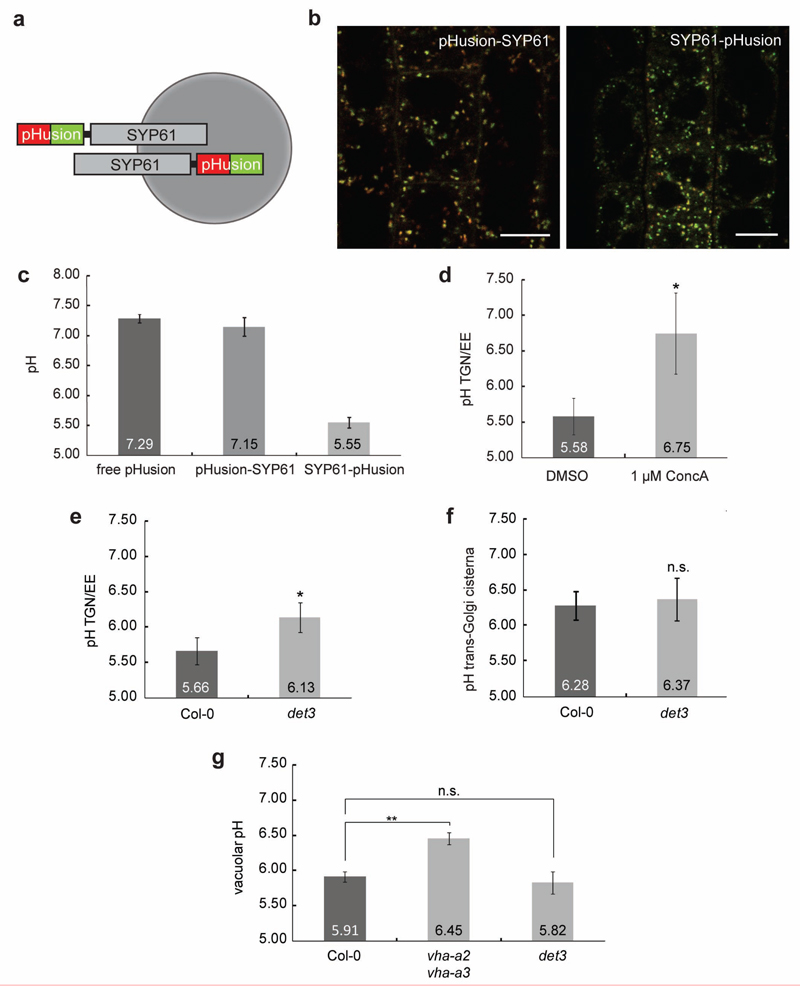
To investigate whether impairment of V-ATPase activity changes the pH of the TGN/EE and the Golgi we fused the ratiometric pH-sensor pHusion18 to the TGN/EE marker SYNTAXIN OF PLANTS61 (SYP61) that co-localizes with VHA-a1 in the TGN/EE19 (Fig. 1a and Supplementary Fig. 1). Similar constructs have been used to determine the localization of Syntaxin1a in mammalian cells20 and thus, we generated stable transgenic lines expressing SYP61-pHusion and pHusion-SYP61. Fluorescence was detected in root meristem epidermal cells (Fig. 1b) and in vivo calibration of SYP61-pHusion, pHusion-SYP61 and cytosolic pHusion (Supplementary Fig. 1) resulted in sigmoidal calibration curves. pH values of 5.6 for SYP61-pHusion and pH 7.2 for both pHusion-SYP61 and cytosolic pHusion (Fig. 1c) corroborated that SYP61-pHusion was targeted to the TGN/EE lumen and was suitable for in vivo pH measurements in this compartment. V-ATPase activity inhibition by 3-h ConcA treatment raised the pH in the TGN/EE from 5.58 to 6.75 (Fig. 1d). In contrast, the pH in the TGN/EE of det3 was 6.13 (Fig. 1e). To check whether V-ATPase activity also determines the pH in trans-Golgi cisternae, we linked pHusion C-terminally to rat sialyltransferase (ST)21. For both, wild type and det3, a pH-value of 6.3 was obtained (Fig. 1f and Supplementary Fig. 1). Similarly, no significant changes in vacuolar pH were detected in the det3 mutant (pH 5.82) compared to the wild type (pH 5.92), whereas in vha-a2 vha-a3 double mutant, that lacks tonoplast V-ATPase, the vacuolar pH increased to 6.45 (Fig.1g and Supplementary Fig. 1). To assess the ultrastructure of Golgi-stacks and TGN/EE in the det3 mutant, we used transmission electron microscopy (TEM) sections from roots of light-grown seedlings. In contrast to what has been previously reported for ConcA-treated cells6, the ultrastructure of Golgi stacks and TGN/EE in det3 did not differ significantly from the wild type (Supplementary Fig. 2). In agreement with the TEM data, confocal microscopy-analysis of the TGN/EE-marker SYP61 and trans-Golgi marker ST revealed no differences in size, number and brefeldinA (BFA)-responsiveness22 of the compartments labelled by the respective markers (Supplementary Fig. 2).
Figure 1. pH in the TGN/EE of det3 is increased.
a, The ratiometric pH sensor pHusion fused N-terminally (pHusion-SYP61) and C-terminally (SYP61-pHusion) to SYP61. b, Confocal images of Arabidopsis plants stably expressing N- and C-terminal fusions. Scale bars, 10 μm. c, In vivo pH measurements of transgenic Arabidopsis lines expressing pHusion in the cytosol (free pHusion), N-terminally (pHusion-SYP61) or C-terminally linked to SYP61 (SYP61-pHusion). d, In vivo pH measurements of the TGN/EE in wild type (Col-0) cells treated with 1 μM ConcA for 3 h or equal amounts of DMSO. e, In vivo pH measurements of the TGN/EE in wild type (Col-0) and det3. d and e, Values represent averages of three individual experiments. P values (t -test), *, P < 0.05. f, In vivo pH measurements of the trans-Golgi cisterna in wild type (Col-0) and det3. c and g, Values represent averages of three individual experiments. P values (t -test), not significant (n.s.). g, Vacuolar pH measurements in wild type (Col-0), vha-a2 vha-a3 and det3 epidermal and cortex cells. Values represent averages of three individual experiments P values (t -test), **, P < 0.01, relative to the respective controls, Col-0 for vha-a2 vha-a3 and vha-a2 vha-a3 for det3. 6-day-old seedlings were used for all measurements. Error bars indicate S.D.
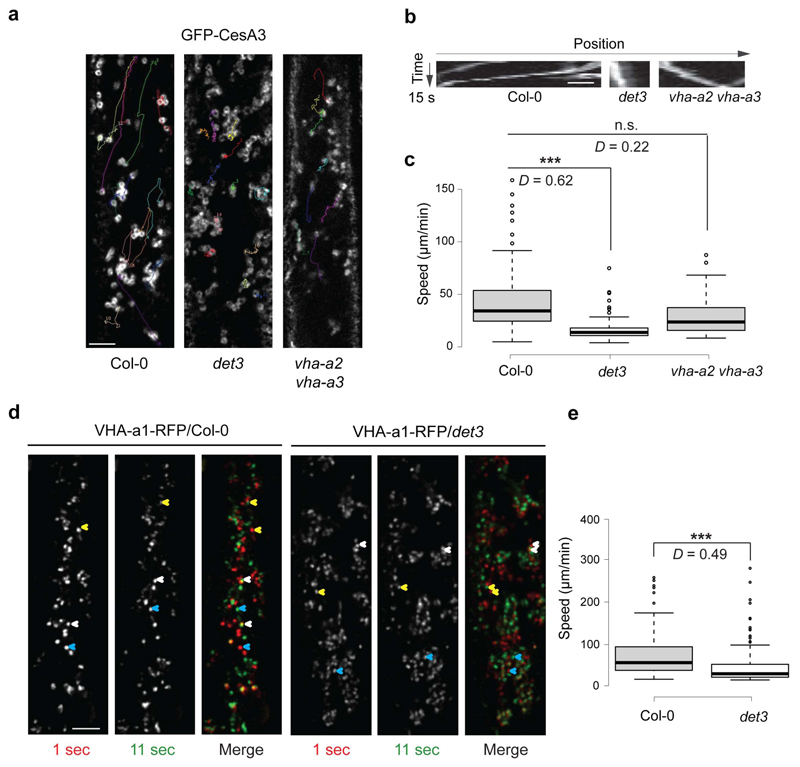
To investigate whether the pH changes of the TGN/EEs in det3 affected the behaviour of the Golgi and TGN/EEs in vivo, we examined the motility of GFP-CesA3, that labels Golgi bodies and partially TGN/EEs16,17 and of the TGN/EE marker VHA-al-RFP1 in the det3 mutant. Live-cell imaging and quantitative analyses indicted that the Golgi and TGN/EE compartments moved more slowly in epidermal cells of both etiolated hypocotyls and root meristems of light-grown det3 plants than in those of the wild type (Fig. 2a-e and Supplementary Fig. 3). In contrast, the motility of the GFP-CesA3 cytoplasmic foci was unaffected in vha-a2 vha-a3 double mutant cells (Fig. 2a-c).
Figure 2. Reduced Golgi and TGN/EE motilities in det3.
a, Reduced speed of GFP-CesA3, which labels Golgi and partly TGN/EE, at cytosolic focal planes in hypocotyl cell of 3-day-old etiolated det3, wild type (Col-0) and vha-a2 vha-a3 seedlings. Only compartments with clear donghnut-like shapes (indicative of Golgi14) were measured. Movement is depicted as lines in a single frame that follow the GFP-CesA3 movement over time as estimated from time average images. b, Kymograph traces of the typically GFP-CesA3 movement in the genotypes and treatment from a. c, Box plots displaying the speed of the Golgi-located GFP-CesA3 in the genotypes from a. Golgi-located GFP-CesA3 in det3 are significantly slower than in the wild type control and vha-a2 vha-a3 cells. Estimates are done on cells from five independent seedlings. Golgi per seedling (n > 25). ***P-value < 0.001; D value from Kolmogorov-Smirnov tests. d, Reduced speed of VHA-a1-RFP, that labels TGN/EE in 3-day-old etiolated det3 hypocotyls when compared to the wild type control. VHA-a1-RFP movement is depicted as changes in VHA-a1-RFP location between two consecutive confocal images (10-sec interval). In merged image, VHA-a1-RFP marked in red and green represents T= 1 sec (first frame) and T= 11 sec (second frame), respectively. Note the differences in distance a typical VHA-a1-RFP signal travelled between the consecutive images in wild type and det3 (exemplified for three VHA-a1-RFP particles by yellow, red, and blue arrowheads). e, Box plots displaying the VHA-a1-RFP speed in the genotypes and treatments from d. VHA-a1-RFP compartments in det3 have significantly reduced motilities as compared to wild type control cells. Estimates were done on cells from five independent seedlings. TGN/EE per seedling (n > 20). ***P-value < 0.001; D value from Kolmogorov-Smirnov tests. Two-tailed Wilcoxon rank sum test (P-value < 0.001) was also performed. Scale bars, 10 μm.
BR signalling is impaired in det3
To verify that det3 had reduced responses to BRs9,10, we evaluated growth of hypocotyl and primary root of dark- and light-grown det3 seedlings, respectively, in the presence of increasing concentrations of brassinolide (BL) (Supplementary Fig. 4). To avoid the reported growth-inhibiting effect of nitrate8, the det3 seedlings were grown on solid medium without salts. Both hypocotyl and root lengths of det3 were partially insensitive to the BL application. We also monitored the phosphorylation status of the transcription factor BRI1 EMS-SUPPRESSOR1 (BES1), routinely used as readout for BR signalling activation23, and found that dephosphorylated BES1 was significantly reduced in det3 mutant when compared to the wild type (Supplementary Fig. 4). In contrast, the vha-a2 vha-a3 mutant showed a slight hypersensitivity to BL and only a minor increase in dephosphorylated BES1 (Supplementary Fig. 4), thus excluding the contribution of tonoplast-localized V-ATPases to the BR insensitive phenotype of det3. Reduced sensitivity to BR was also observed when wild type seedlings were grown in the presence of ConcA (Supplementary Fig. 4). Thus, the decreased responses to BRs in det3 result from the impaired V-ATPase function in the TGN/EE.
Defective endocytic trafficking in det3 does not cause BR insensitivity
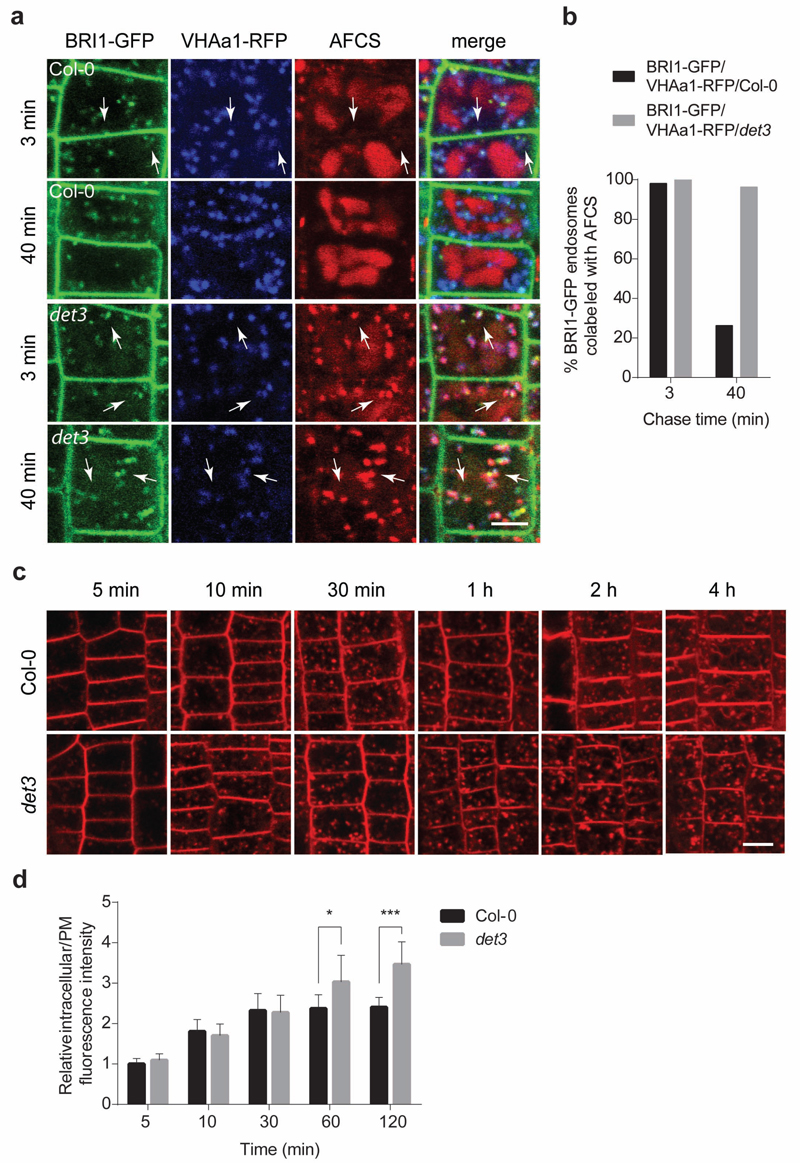
BRI1-ligand complexes undergo constitutive endocytosis1,2,12,24, that can be visualized by the bioactive fluorescent BR, Alexa Fluor 647-catasterone (AFCS)13. Therefore, we investigated whether the endocytic trafficking of BRI1-BR complexes is impaired in det3. In root epidermal cells of det3, AFCS strikingly accumulated in the TGN/EEs, marked by the VHA-a1-RFP1, together with BRI1-GFP25 (Fig. 3a,b), similarly to the internalization in the presence of ConcA13. In contrast to the wild type and the vha-a2 vha-a3 double mutant, in which AFCS largely accumulated in the vacuole (Supplementary Fig. 5), in det3 AFCS occurred mainly in the TGN/EEs, with only a minor accumulation in the vacuole, even after a 40-min chase. Together these data indicate that the endocytic trafficking from TGN/EE to the vacuole is delayed in det3. To address the question whether internalization of plasma membrane material in general is affected in the det3 mutant, we quantified the uptake of the fluorescent styryl dye FM4-64, routinely used as endocytic tracer in plants26, in root epidermal cells. No significant differences between wild type and det3 were observed when the FM4-64 uptake was quantified at early time points (Fig. 3c,d). Whereas a 4-h incubation with FM4-64 resulted in a clear tonoplast labelling in the wild type, the tonoplast staining in det3 was only faint(Fig. 3c,d), indicating that the endocytic trafficking from TGN/EE to the vacuole in det3 is delayed, contrary to the described complete block of FM4-64 internalization in TGN/EE by ConcA1.
Figure 3. Endocytic trafficking from TGN/EE to the vacuole is delayed in det3.
a, Pulse-chase AFCS uptake experiments in BRI1-GFP/VHAa1-RFP/Col-0 and BRI1-GFP/VHAa1-RFP/det3 Arabidopsis lines. Seedlings were incubated in AFCS (20 μM) for 20 min followed by either a f 3-min or 40-min chase as indicated. b, Quantification of the percentage of VHAa1-RFP positive endosomes labelled by both BRI1-GFP and AFCS in a. c, FM4-64 (5 μM) uptake for 5 min, 10 min, 30 min, 1 h, 2 h, and 4 h. Arabidopsis root epidermal cells of 5-day-old wild type (Col-0) and det3 seedlings were imaged immediately after washing. d, Maximal intracellular fluorescence intensity normalized to the maximal fluorescence intensity at the plasma membrane (PM) in root epidermal cells of wild type (Col-0) and det3 after incubation with FM4-64 as in c. At least 15 cells from three roots were calculated for each time point. No significant differences were observed for 5 min, 10 min, and 30 min (t -test). P values (t -test), * P < 0.05, *** P < 0.001. Error bars indicate S.D. Scale bars, 5 μm.
As reported earlier13, and in contrast to det3 and to ConcA-grown plants (Supplementary Fig. 4), short ConcA treatments (2 μM, 2 h) had no effect on BR responses in roots, as evaluated by the BES1 dephosphorylation assay (Supplementary Fig. 5). Hence, although endocytic trafficking between the TGN/EE and the vacuole is affected in det3, this phenotype does not explain the reduced BR sensitivity of the mutant.
Secretion defects of BRI1 contribute to the impaired BR signalling in det3
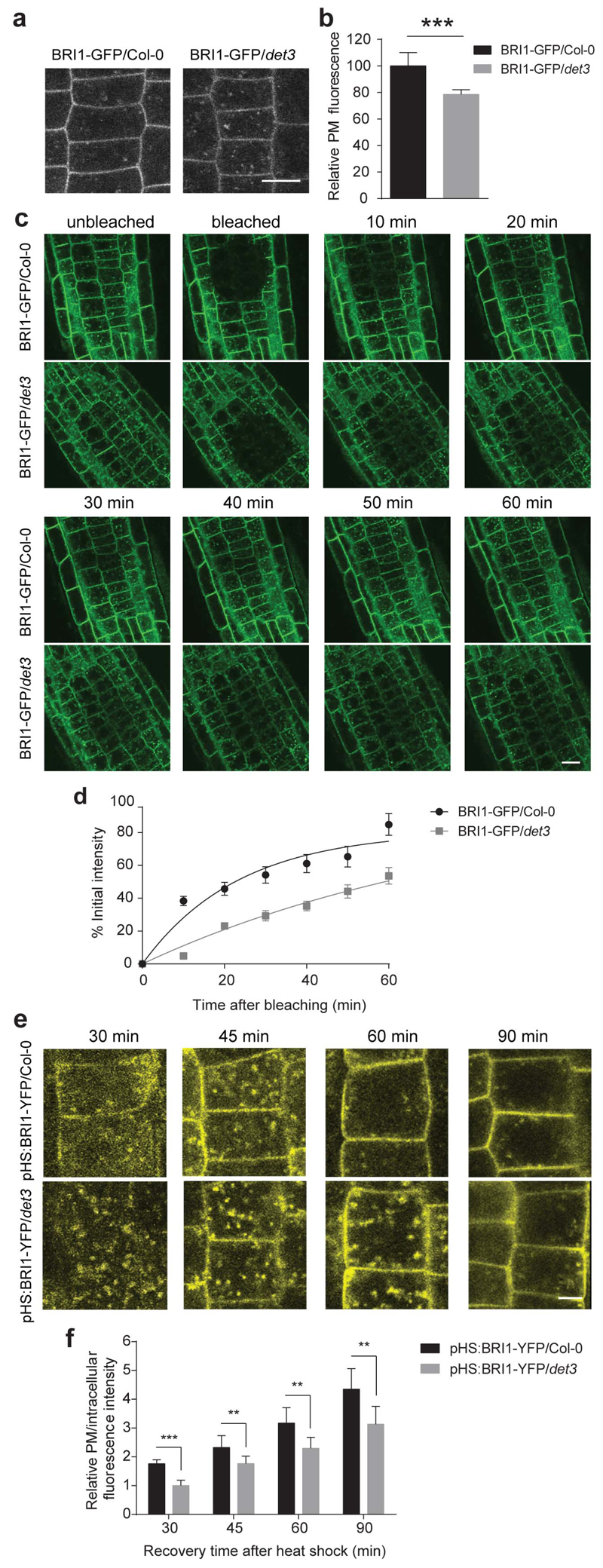
Plasma membrane-localized BRI1 accounts for most of the BR signalling13. To unravel the BR insensitive phenotype of the det3 mutant, we introduced the pBRI1::BRI1-GFP construct12 into det3. No significant differences in BRI1 expression and protein levels were observed between the wild type and the det3 (Supplementary Fig. 6). However, the relative plasma membrane BRI1-GFP fluorescence in root and hypocotyl epidermal cells was approximately 20% reduced in det3 (Fig. 4a,b and Supplementary Fig. 6), suggesting that the BR insensitivity of the mutant might be caused by decreased BRI1 levels in the plasma membrane.
Figure 4. Secretion of BRI1 in det3 is reduced.
a, Confocal images of 5-day-old BRI1-GFP/Col-0 and BRI1-GFP/det3 root epidermal cells. Scale bar, 5 μm. b, Quantification of relative plasma membrane (PM) fluorescence intensity of BRI1-GFP/Col-0 and BRI1-GFP/det3 as shown in a (30 cells from five roots were measured). Plasma membrane fluorescence was normalized to background fluorescence for each measurement. P values (t -test), *** P< 0.001 relative to the respective control. c, FRAP analysis of BRI1-GFP/Col-0 and BRI1-GFP/det3 plasma membrane BRI1 recovery rate. Consistent ROI was selected for individual roots, followed by photobleaching to significantly reduce the target area fluorescence. Root epidermal cell fluorescence was recorded before bleaching and at different time points after bleaching. Scale bar, 10 μm. d, The fluorescence recovery was calculated on cells with totally bleached whole plasma membrane fluorescence. Plasma membrane fluorescence before and immediately after bleaching was set as 100 % and 0 %, respectively. Each recovery value was normalized to fluorescence value of the unbleached region (at least 15 cells from three roots were measured). e, In vivo analysis of heat shock-induced BRI1-YFP exocytosis. YFP signal in 5-day-old pHS:BRI1-YFP/Col-0 and pHS:BRI1-YFP/det3 root epidermal cells chased at 30 min, 45 min, 60 min, and 90 min after 1-h 37°C induction. Scale bar, 5 μm. f, Relative plasma membrane to intracellular fluorescence intensity values calculated by fluorescence intensities measured using ImageJ. ROI was kept constant for each measurement (at least 15 cells from three roots were measured). Error bars indicate S.D.
To assess whether BRI1 trafficking to the plasma membrane is affected in det3, we employed fluorescence recovery after photobleaching (FRAP) in root epidermal cells. The recovery of the plasma membrane BRI1-GFP fluorescence in det3 was remarkably slower than that of the wild type (Fig. 4c,d), suggesting that the BRI1 exocytosis was impaired. As a further validation, we introduced the heat-shock inducible BRI1-YFP12 into det3, ensuring that the expression levels of BRI1-YFP after the heat shock were the same in wild type and det3 (Supplementary Fig. 6) and monitored the YFP fluorescence in a time course manner after the heat shock. After a 30-min recovery, most of the BRI1-YFP signal was intracellular in det3, whereas a considerable amount of BRI1-YFP was already located in the plasma membrane of the wild type (Fig. 4e). Quantitative analyses after 90 min revealed that the plasma membrane BRI1-YFP was significantly lower in det3 than that in the wild type (Fig. 4f), indicating that BRI1 secretion was partially inhibited in det3.
BRI1 recycling is defective in det3
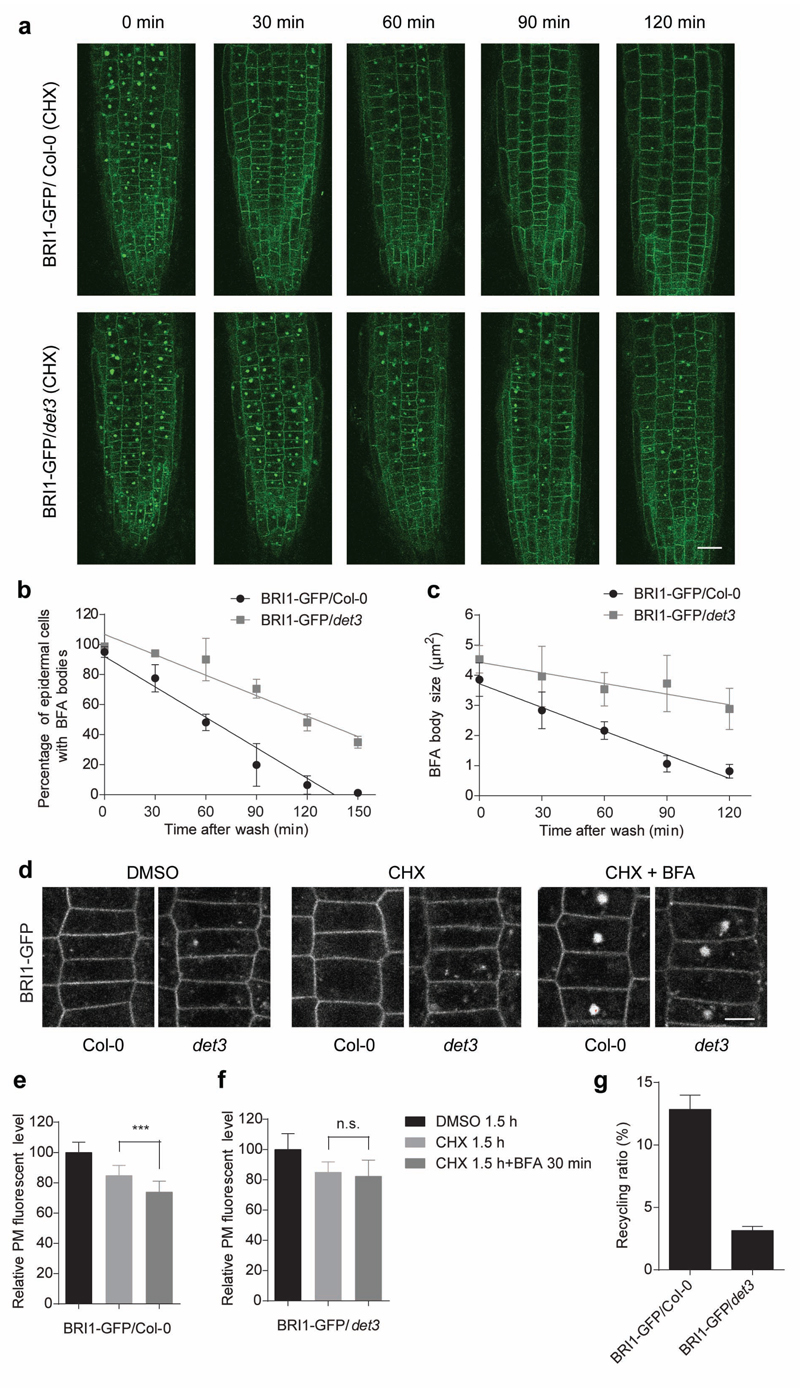
To assess whether recycling contributes to the exocytosis defects of det3 we performed washout experiments after BFA22 application (50 μM, 30 min) in the presence of cycloheximide (CHX) (50 μM) in det3/BRI1-GFP and wild type roots (Fig. 5a). The number of BFA body-containing cells and the BFA body sizes were scored at different time points after washing. Almost all BRI1-GFP was relocated from the BFA body after 90 min of BFA washout in wild type cells but not in det3 cells even after 150 min (Fig. 5a,b). Quantitative analyses also revealed that the BFA compartment size decreased more slowly after BFA removal in the det3 mutant than in the wild type (Fig. 5c).
Figure 5. BRI1 recycling from TGN/EE to plasma membrane is reduced in det3.
a, BRI1-GFP in roots of 5-day-old wild type (Col-0) and det3 seedlings pretreated with cycloheximide (CHX) (50 μM) for 1 h, followed by treatment for 30 min with CHX (50 μM) plus brefeldinA (BFA) (50 μM). Seedlings were washed in CHX (50 μM) and mounted for imaging at 0 min, 30 min, 60 min, 90 min, and 120 min after BFA washout. Scale bar, 20 μm. b and c, BRI1-GFP relocalization from the BFA bodies to the plasma membrane was quantified by either as percentage of cells with BFA body in b or BFA body size in c. d, BRI1-GFP fluorescence of 5-day-old wild type (Col-0) and det3 root epidermal cells treated separately with DMSO (1 %) and CHX (50 μM) for 1.5 h and compared with seedlings treated with BFA (50 μM) for 30 min in the presence of CHX (50 μM) for 1.5 h. Scale bar, 20 μm. e and f, Quantification of relative plasma membrane fluorescence intensities of BRI1-GFP in e and BRI1-GFP/det3 in f of the root epidermal cells represented in d (for each treatment, at least 15 cells from three roots were measured). P values (t -test), ***, P< 0.001 relative to the respective control (CHX). g, Recycling of BRI1-GFP protein in BRI1-GFP/Col-0 and BRI1-GFP/det3 (at least 15 cells from three roots were measured). Error bars indicate S.D.
To evaluate the contribution of recycling to the plasma membrane pool of BRI1 in det3, we measured the plasma membrane levels of BRI1-GFP in the presence of CHX and after a combined BFA and CHX treatment (Fig. 5d-f) and calculated the recycling ratio as the relative reduction in plasma membrane fluorescence intensity after BFA application (Fig. 5g). The CHX treatment decreased the plasma membrane BRI1 levels in both wild type and det3 epidermal root cells with approximately 10% due to inhibited de novo protein synthesis. Application of BFA in the presence of CHX additionally reduced the plasma membrane BRI1 pool, thus reflecting the recycling contribution (Fig. 5d-f). Interestingly, this reduction was lower in det3 than the wild type (recycling ratios of 4 % and 13 %, respectively; Fig. 5g).
Previously BRI1 endocytosis and degradation had been shown to be independent of its ligand12,24. Consistently, application of BL in the presence of CHX had no impact on BRI1-GFP fluorescence intensity at the plasma membrane in root epidermal cells of wild type and det3 (Supplementary Fig. 7), germinated with or without brassinazole (BRZ), a BR biosynthesis inhibitor27. The recycling ratio was also not affected by the depletion of BRs in either genotypes (Supplementary Fig. 8). Interestingly, the BL application was able to release some constrained BRI1 from BFA bodies to the plasma membrane in wild type cells only when grown in the presence of BRZ, indicating the existence of a BFA-insensitive BRI1 recycling route (Supplementary Fig. 9). Hence, we conclude that BRI1 recycling is impaired in det3, thus contributing to its reduced plasma membrane pool.
The cellulose-related defects in det3 are due to reduced exocytosis of cellulose synthase
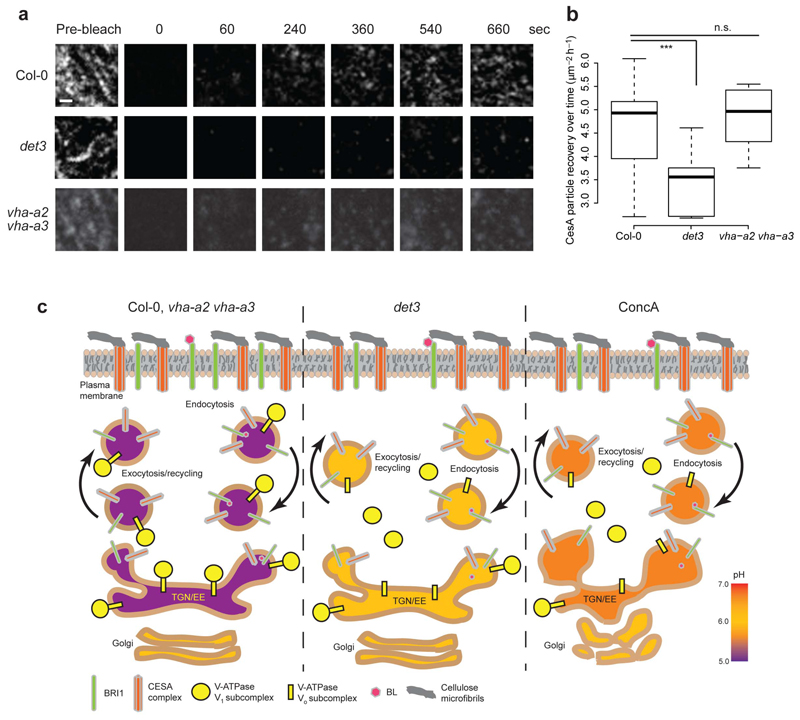
Cellulose levels are reduced in det3 (Supplementary Fig. 10), and mutant seedlings are hypersensitive to cellulose biosynthesis inhibitors8,11. Similarly, seedlings grown on ConcA also contained lower levels of cellulose than the wild type and the vha-a2vha-a3 double mutant (Supplementary Fig. 10). In contrast, other cell wall components were seemingly not affected in det3 (Supplementary Fig. 10). Cellulose is synthesized by plasma membrane-located CesA proteins and the speed of CesAs has been used as a proxy for complex activity14. We analysed the behaviour of GFP-CesA3 in det328, and found that the speed of the plasma membrane-located GFP-CesA3 was similar to wild type in hypocotyl cells of det3 and in vha-a2 vha-a3, and in seedlings treated with ConcA (Supplementary Fig. 11). Similarly, no differences were observed in cellulose microfibril organization among genotypes and treatments as assessed with the cellulose binding dye Pontamine Fast Scarlet 4B29 (Supplementary Fig. 11). These data suggest that the plasma membrane-based activity of the CesA proteins is not impaired in the det3 mutant. To test whether the CesAs trafficking is affected in det3, consistently with the observations of BRI1, we performed FRAP experiments of the plasma membrane plane of GFP-CesA3-expressing hypocotyl cells. The GFP-CesA3 repopulated the plasma membrane considerably slower in the det3 mutant than in the wild type and the vha-a2 vha-a3 double mutant (Fig. 6a,b). Notably, the CesA density at the plasma membrane was similar in det3, wild type and, vha-a2 vha-a3 double mutant (Supplementary Fig. 11). These data are consistent with the defects observed in for BRI1 secretion and suggest that the cellulose deficiency in det3 may be due to a reduced secretion, rather than activity, of the CesA complexes.
Figure 6. Delivery of CesA complexes to the plasma membrane is impaired in det3.
a, CesA delivery events after photobleaching in 3-day-old etiolated hypocotyl cells. The GFP channel was photobleached and the CesA delivery rate calculated44. Scale bar, 1 μm. b, Box plot showing CesA recovery rates in wild type (Col-0), det3, and vha-a2 vha-a3 mutant seedlings. P values (t -test), *** P< 0.001 (n = 6 cells for each condition). c, Schematic model for the impact of the DET3 mutation on endomembranes trafficking in plant cells. In yeast a functional VMA5 (VHA-C) subunit is required for the pump assembly48. In wild type and the vha-a2 vha-a3 mutant plants the pH of the TGN/EE is acidic (5.6), and Golgi and TGN/EE are fully functional. In det3 mutant the pH of the TGN/EE is less acidic (6.1) but the morphology is not affected. In contrast, in plant cells treated with ConcA the pH in the TGN/EE is increased to 6.5 and is causing changes in Golgi and TGN/EE morphology. The change in pH leads to decreased Golgi and TGN/EE motility and respectively reduced secretion of BRI1 and the CesA These defects result in an impaired ability of the plant to respond to BRs and to produce cellulose.
Discussion
Here, we show conclusively that the reduced V-ATPase activity in the det3 mutant affects the steady-state pH in the TGN/EE but not in the Golgi stack or in the vacuole. Surprisingly, this deficiency led to reduced motilities of both, Golgi and TGN/EE compartments. Whereas V-ATPases are not located in the Golgi stack, the reduced ability of the TGN/EE to secrete molecules might cause a feedback loop, leading to changes in the Golgi lumen. For example, monensin treatment of sycamore maple (Acer pseudoplatanus) suspension cells resulted in an initial TGN swelling and, subsequently, of the Golgi cisternae30. The reduced Golgi and TGN/EE motilities in det3 and in ConcA-treated cells might also be related to the ability of V-ATPases to interact with actin filaments31 or to changes in the engagement of myosin32 or other proteins associated with this function, as shown for small GTPases in animal cells33.
Endosomal pH influences the dissociation and degradation of internalized ligands and signalling duration34,35. We found that in det3 the internalization of the BR-bound BRI1 from the plasma membrane to the TGN/EE is unaffected, but that the trafficking to the vacuole is delayed.
Consistent with the reduced BR sensitivity of det3 and wild type seedlings grown on ConcA, we discovered that the amount of BRI1 receptor at the plasma membrane was lower in det3 than in the wild type. Indeed, swollen TGN/EE accumulated BRI1 after ConcA treatment2, suggesting that BRI1 secretion might be impaired in det3, as confirmed by FRAP experiments and the inducible BRI1 construct.
Besides secretion, recycling also contributes to the plasma membrane pool of BRI1. BFA experiments revealed a delay in BRI1 recycling in det3. The effect was the same in plants grown with or without the BR biosynthesis inhibitor. Recycling of plasma membrane receptors in animal cells requires the dissociation of ligands in the acidic environment of the EEs. BR binding to BRI1 takes place in the acidic plant apoplast. In addition, the BL-induced interaction of BRI1 with its coreceptor, the BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1) is promoted by low pH36. In contrast to mammalian cells, the pH in the plant TGN/EE is lower than that of the late endosomal compartments37–39, but does not differ much from that in the apoplast. As the acidic pH typically favours the BRI1-BR-BAK1 complex, the expected pH-dependent BR dissociation from BRI1 in TGN/EE might not occur, and we most likely monitored the constitutive cycling of nonactive BRI1 between the plasma membrane and the endosome (Fig. 6c). Overall, our data suggest that plants might differ from animals in terms of receptor-ligand dissociation and recycling.
In addition to the BR-related phenotype, det3 also contained less cellulose8, as revealed by the reduced CesA secretion to the plasma membrane and consistent with the BRI1 trafficking defects (Fig. 6c). Nevertheless, the CesA density at the plasma membrane was similar to that of the wild type. CesA and other cell wall components that are synthesised in the Golgi might plausibly be transported to the cell surface via different routes, as illustrated by the TGN-localized protein ECHIDNA (ECH), which affects secretion of cell wall material, but not that of PIN-FORMED2 (PIN2) and PIN340,41. If so, the ratio of cellulose-to cell wall matrix polymers would be changed in det3 as indeed observed (Supplementary Fig. 10). In addition, because the CesA density is the same in det3 and wild type cells, also internalization of the proteins might be assumed to be affected, and an increase in stalled CesA awaiting internalization might be anticipated Closer inspection of slowly moving CesA in the plasma membrane revealed that det3 cells contain about three fold as many slow moving CesA(below 150 nm/min) (Supplementary Fig. 11) supporting a delay in internalization.
Whereas it is clear that the BRI1 and CesA secretion to the plasma membrane is impaired due to defects in the endomembrane pH, the reason for these defects is still unknown. Changes in the endomembrane pH may influence cargo processing such as glycosylation‥ Indeed, mis-glycosylation of the CesA-associated endo-glucanase Korrigan caused defects in the complex trafficking and led to cellulose deficiency42. The altered pH might also affect recruitment and tethering of the trafficking machinery on the cytosolic side of the endomembrane. Such regulation has, for example, been shown for the small GTPases Arf6 and ARNO in animal cells43. Although the exact process that underpins the secretion defects remains to be determined, our data propose an explanation the observed phenotypic defects in the V-ATPase-related mutant det3 (Fig. 6c).
Methods
Plant materials and growth conditions
Arabidopsis thaliana (L.) Heyhn. (Columbia accession, Col-0) seeds were stratified for 1 to 2 days at 4 °C to synchronize germination and grown vertically on half-strength Murashige and Skoog (1/2 MS) (Duchefa) containing 1% (w/v) sucrose and 1% agar at 22°C in a 16-h/8-h light-dark cycle for different periods of time depending on the experiments. For the BR growth assays in the dark or the light, seeds were plated on medium containing 1% phytagar, 1% sucrose, adjusted to pH 5.8 with 20 mM MES, pulsed with light for 4 h before transfer to darkness or kept in the light for 5 days. Etiolated seedlings were grown as described44. ConcanamycinA (ConcA), brassinolide (BL), or brassinazole (BRZ) were dissolved in the medium at the concentrations indicated in the figures. BRI1-GFP/det3, GFP-CesA3/det3, VHA-a1-RFP/det3, and pHS:BRI1-YFP/det3 were generated by crossing det3 (ref. 8) with endogenous BRI1-GFP12, GFP-CesA3 (ref. 28), VHA-a1-RFP1 and pHS:BRI1-YFP12, respectively. AFCS uptake was performed on det3 seedlings crossed with the BRI1-GFP overexpressing (OE) line25 and with VHA-a1-RFP. The vha-a2 vha-a3 double mutant was described previously7.
Generation of constructs
Entire cloning procedures and manipulations of DNA were done according to standard and established protocols. Specifications of each primer used during the cloning procedures are listed in Supplementary Table 1. All coding sequences were amplified via PCR from plasmid DNA or first-strand cDNA. Vectors were opened with restriction enzymes resembling the fragments. Escherichia coli strain DH5-α was used for all procedures. Overexpression of all constructs generated was driven by the p16 ribosomal promoter45 out of a pTkan vector backbone46,47. The p16 promoter sequence was amplified via PCR out of a pP16Tkan plasmid (kind gift of Dr. Christoph Neubert) with primers P16-KpnI-fwd and P16-KpnI-rev, introducing two KpnI restriction sites. The P16 fragment was subsequently ligated into the pTkan vector cut with KpnI, and its correct orientation was confirmed via restriction digest. The P16Tkan vector was used as basic vector for all further generated constructs.
For the SYP61-pHusion construct, the Arabidopsis SYP61-coding sequence (AT1G28490) was amplified from the Arabidopsis cDNA with the primers SYP61-BamHI-fwd/SYP61-HindIII [noSTOP]-rev primers, removing the Stop codon. pHusion was amplified from 35S:pHusion plasmid18 with pHusion-HindIII-fwd and pHusion-SalI-rev primers. Fragments were sub-cloned into pJETblunt1.2 (Fermentas) for sequencing and multiplication. After cutting with the respective restriction enzymes, fragments were ligated into the p16Tkan vector via the BamHI/SalI sites. The pHusion-SYP61 construct was generated by amplifying SYP61 with the primers SYP61-HindIII-fwd and SYP61-SalI-rev as well as pHusion (pHusion-BamHI-fwd/pHusion [noSTOP]-HindIII-rev). Both fragments were removed from pJETblunt1.2 intermediate vector after sequencing with the respective restriction enzymes and inserted into p16TKan cut in the same manner. For ST-pHusion constructs, sialyltransferase coding sequence was amplified with ST-AatII-fwd and ST-PvuI-rev primers, sub-cloned into pJET blunt1.2 and removed with respective restriction enzymes. Together with pHusion, which was amplified with primers pHusion-PvuI-fwd and pHusion-SalI-rev and sub-cloned identically, ST was ligated into p16Tkan, pre-cut with AatII and SalI.
In vivo pH measurements in the TGN/EE
pH was determined in root cells of 6-day-old T2 and T3 of SYP61-pHusion and pHusion-SYP61 seedlings. The homozygous lines expressing the cytoplasmic pHusion have been described previously18. A Leica SP5 II confocal laser-scanning microscope equipped with two Hybrid Detectors was used for imaging. Images were obtained with a HCX PL APO λ blue 63.0 × 1.20 UV water immersion objective. pHusion was excited in single scans from eGFP with a VIS-Argon laser at 488 nm and from mRFP at 561 nm with a VIS-DPSS561 laser. Emission was detected for eGFP and mRFP between 490 and 545 nm and between 600 to 670 nm, respectively. Images for free cytosolic pHusion were obtained sequentially due to spectral bleed-through. Background and oversaturation were excluded by thresholding with ImageJ version 1.49d (National Institute of Health). Average intensities for each channel were obtained and eGFP/ mRFP ratios were calculated. In vivo calibration was achieved by incubating seedlings for 15 min in pH equilibration buffers of 50 mM MES-Bis-tris-propane (BTP) (pH 5.2 to 6.4) or 50 mM Hepes-BTP and 50 mM ammonium acetate. pH equilibration buffer (pH=4.8) consisted of 50 mM citrate buffer (22 mM citric acid and 27 mM tri-sodium citrate; adjusted with HCl) and 50 mM ammonium acetate. A sigmoid calibration curve was obtained by assigning ratios to the corresponding pH values in a Boltzmann fit by means of the Origin Pro 9.1G. pH values of all lines used were calculated equally.
In vivo vacuolar pH measurements
Vacuolar pH measurements and BCECF calibration were performed as previously described7 with slight modifications. Seedlings were grown for 6 days (1/2 MS, pH5.8, 1% phytoagar) and stained in respective liquid medium containing 10 μM BCECFAM[(2',7'-Bis-(2-carboxyethyl)-5(6)-carboxyfluorescein]-(acetoxymethylester) (Molecular Probes, Invitrogen) in the presence of 0.02 % Pluronic F-127 (Molecular Probes, Invitrogen) for 1h in the dark at 22°C, and washed twice for 5min with medium. Imaging was done on a Leica SP5 Ilusing a HCX PL APO CS 20 × 0.70 IMM UV objective. BCECF was exited sequentially at 458 nm and 488 nm. Emission was detected between 510 and 550 nm. Average fluorescence intensity values were obtained using ImageJ version 1.49d (National Institute of Health). Calibration and calculation of a calibration curve were done as for the pH measurements in the TGN/EE.
Supplementary Material
Acknowledgements
We thank Yanhai Yin (Iowa State University, Ames, USA) for providing the anti-BES1 antibody, Niko Geldner (Univeritè de Lausanne, Switzerland) for sharing pBRI1:BRI1GFP and pHS:BRI1-YFP lines, Daniël Van Damme (VIB-Ghent University, Belgium) for advice in microscopy, and Martine De Cock (Ghent University, Belgium) for help in preparing the manuscript. This work was supported by the Marie-Curie Initial Training Network ‘BRAVISSIMO’ (grant no. PITN-GA-2008-215118) to E.R., the Odysseus program of the Research Foundation-Flanders to J.F., the Max-Planck Gesellschaft to A.D., Y.Z., L.N., and S.P., the European Union Seventh Framework Programme (FP7 2007-2013) under Grant Agreement 263916 (WallTrac, Marie Curie Initial Training Network) to P.K. and the Deutsche Forschungsgemeinschaft (SFB1101 and TPA02) to K.S‥ Y.L. is indebted to the Belgian Science Policy Office (BELSPO) for a postdoctoral fellowship.
Footnotes
Author contributions
Y.L., A.D., S.S., N.G.I., L.N., J.F, K.S., E.R., and S.P. conceived the study and designed the experiments. Y.L. performed all BR-related work, A.D., Y.Z., and P.K. performed all CesA-related work, S.S. constructed the pH sensor and did all pH measurements, N.G.I. did the AFCS uptake, Y.L. A.D., Y.Z., N.G.I., S.D.R., and I.V.H. generated materials, Y.-D. S. and K.S. did the TEM, Y.L., A.D., Y.Z., N.G.I., L.N., P.K., E.M., and V.B. did imaging, J.W. performed the chemical synthesis of AFCS, Y.L. A.D., N.G.I., S.D.R., S.P., K.S., and E.R. wrote the manuscript. All authors commented on the results and the manuscript.
Additional information
Supplementary information is available on line.
Competing interests
The authors declare no competing financial interests
References
- 1.Dettmer J, Hong-Hermesdorf A, Stierhof Y-D, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viotti C, et al. Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell. 2010;22:1344–1357. doi: 10.1105/tpc.109.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishi T, Forgac M. The vacuolar (H+)-ATPases -- nature’s most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 4.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C, Chang A. pH-dependent cargo sorting from the Golgi. J Biol Chem. 2011;286:10058–10065. doi: 10.1074/jbc.M110.197889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dettmer J, et al. Essential role of the V-ATPase in male gametophyte development. Plant J. 2005;41:117–124. doi: 10.1111/j.1365-313X.2004.02282.x. [DOI] [PubMed] [Google Scholar]
- 7.Krebs M, et al. Arabidopsis V-ATPase activity at the tonoplast is required for efficient nutrient storage but not for sodium accumulation. Proc Natl Acad Sci USA. 2010;107:3251–3256. doi: 10.1073/pnas.0913035107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brüx A, et al. Reduced V-ATPase activity in the trans-Golgi network causes oxylipin-dependent hypocotyl growth inhibition in Arabidopsis. Plant Cell. 2008;20:1088–1100. doi: 10.1105/tpc.108.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schumacher K, et al. The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes Dev. 1999;13:3259–3270. doi: 10.1101/gad.13.24.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabrera y Poch HL, Peto CA, Chory J. A mutation in the Arabidopsis DET3 gene uncouples photoregulated leaf development from gene expression and chloroplast biogenesis. Plant J. 1993;4:671–682. [Google Scholar]
- 11.Caño-Delgado A, Penfield S, Smith C, Catley M, Bevan M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 2003;34:351–362. doi: 10.1046/j.1365-313x.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- 12.Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007;21:1598–1602. doi: 10.1101/gad.1561307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irani NG, et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol. 2012;8:583–589. doi: 10.1038/nchembio.958. [DOI] [PubMed] [Google Scholar]
- 14.Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- 15.McFarlane HE, Döring A, Persson S. The cell biology of cellulose synthesis. Annu Rev Plant Biol. 2014;65:69–94. doi: 10.1146/annurev-arplant-050213-040240. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 17.Crowell EF, et al. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21:1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gjetting SK, Ytting CK, Schulz A, Fuglsang AT. Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J Exp Bot. 2012;63:3207–3218. doi: 10.1093/jxb/ers040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell. 2001;12:3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Xu P, Xiao Y, Xiong X, Xu T. Domain requirement for the membrane trafficking and targeting of syntaxin 1A. J Biol Chem. 2006;281:15457–15463. doi: 10.1074/jbc.M513246200. [DOI] [PubMed] [Google Scholar]
- 21.Wee EG-T, Sherrier DJ, Prime TA, Dupree P. Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell. 1998;10:1759–1768. doi: 10.1105/tpc.10.10.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geldner N, Friml J, Stierhof Y-D, Jürgens G, Palme K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature. 2001;413:425–428. doi: 10.1038/35096571. [DOI] [PubMed] [Google Scholar]
- 23.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 24.Russinova E, et al. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrichsen DM, Joazeiro CAP, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1255. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jelínková A, et al. Probing plant membranes with FM dyes: tracking, dragging or blocking? Plant J. 2010;61:883–892. doi: 10.1111/j.1365-313X.2009.04102.x. [DOI] [PubMed] [Google Scholar]
- 27.Asami T, et al. Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 2000;123:93–99. doi: 10.1104/pp.123.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desprez T, et al. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:15572–15577. doi: 10.1073/pnas.0706569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson CT, Carroll A, Akhmetova L, Somerville C. Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 2010;152:787–796. doi: 10.1104/pp.109.150128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang GF, Driouich A, Staehelin LA. Effect of monensin on plant Golgi: re-examination of the monensin-induced changes in cisternal architecture and functional activities of the Golgi apparatus of sycamore suspension-cultured cells. J Cell Sci. 1993;104:819–831. doi: 10.1242/jcs.104.3.819. [DOI] [PubMed] [Google Scholar]
- 31.Ma B, et al. Arabidopsis vacuolar H+-ATPase (V-ATPase) B subunits are involved in actin cytoskeleton remodeling via binding to, bundling, and stabilizing F-actin. J Biol Chem. 2012;287:19008–19017. doi: 10.1074/jbc.M111.281873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sparkes IA. Motoring around the plant cell: insights from plant myosins. Biochem Soc Trans. 2010;38:833–838. doi: 10.1042/BST0380833. [DOI] [PubMed] [Google Scholar]
- 33.Maranda B, et al. Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J Biol Chem. 2001;276:18540–18550. doi: 10.1074/jbc.M011577200. [DOI] [PubMed] [Google Scholar]
- 34.Marshansky V, Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gidon A, et al. Endosomal GPCR signaling turned off by negative feedback actions of PKA and v-ATPase. Nat Chem Biol. 2014;10:707–709. doi: 10.1038/nchembio.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Y, et al. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013;23:1326–1329. doi: 10.1038/cr.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinière A, et al. In vivo intracellular pH measurements in tobacco and Arabidopsis reveal an unexpected pH gradient in the endomembrane system. Plant Cell. 2013;25:4028–4043. doi: 10.1105/tpc.113.116897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen J, et al. Organelle pH in the Arabidopsis endomembrane system. Mol Plant. 2013;6:1419–1437. doi: 10.1093/mp/sst079. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher K. pH in the plant endomembrane system -- an import and export business. Curr Opin Plant Biol. 2014;22:71–76. doi: 10.1016/j.pbi.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Boutté Y, et al. ECHIDNA-mediated post-Golgi trafficking of auxin carriers for differential cell elongation. Proc Natl Acad Sci USA. 2013;110:16259–16264. doi: 10.1073/pnas.1309057110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gendre D, et al. Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell. 2013;25:2633–2646. doi: 10.1105/tpc.113.112482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang JS, et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA. 2008;105:5933–5938. doi: 10.1073/pnas.0800237105. [Err. Proc. Natl. Acad. Sci. USA105, 7893 (2008)]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hurtado-Lorenzo A, et al. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol. 2006;8:124–136. doi: 10.1038/ncb1348. [DOI] [PubMed] [Google Scholar]
- 44.Sampathkumar A, et al. Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 2013;162:675–688. doi: 10.1104/pp.113.215277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlücking K, et al. A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol Plant. 2013;6:1814–1829. doi: 10.1093/mp/sst065. [DOI] [PubMed] [Google Scholar]
- 46.Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 47.Schaaf G, et al. AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J Biol Chem. 2006;281:25532–25540. doi: 10.1074/jbc.M601062200. [DOI] [PubMed] [Google Scholar]
- 48.Ho MN, Hill KJ, Lindorfer MA, Stevens TH. Isolation of vacuolar membrane H+-ATPase-deficient yeast mutants; the VMA5 and VMA4 genes are essential for assembly and activity of the vacuolar H+-ATPase. J Biol Chem. 1993;268:221–227. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.