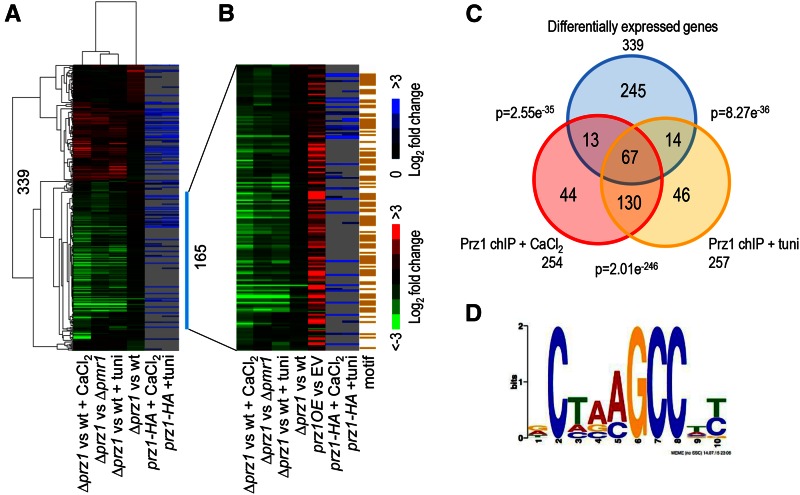
Figure 2.
Identification of Prz1 target genes by transcriptome and ChIP-chip profiling. (A) The heat map shows two-dimensional hierarchical clustering of 339 genes that were differentially expressed by at least twofold in at least one of the microarray experiments. The first four columns of the heat map compare transcriptomes of the following conditions: the ∆prz1 strain and wild type, the ∆prz1 strain and wild type supplemented with 0.15 M CaCl2 for 0.5 hr, the ∆prz1 strain and wild type supplemented with 2.5 µg/ml tunicamycin for 1.5 hr, and the ∆prz1 strain compared to the ∆pmr1 strain. All of the above experiments were performed in rich medium. In the heat map, genes upregulated and downregulated in the ∆prz1 strain relative to the control are indicated in red and green, respectively. The two rightmost columns in the heat map show ChIP-chip analysis of a prz1-HA strain treated with 0.15 M CaCl2 or 2.5 µg/ml tunicamycin for 0.5 and 1.5 hr, respectively. (B) The heat map shows the expression profiles of 165 putative target genes that are positively regulated by Prz1. The first four columns of the heat map match the expression data from A while the fifth column shows the expression profiles of the same target genes upregulated in a prz1+ overexpression strain compared to the empty vector (EV) control. The next two columns in the heat map show ChIP-chip analysis of a prz1-HA strain treated with 0.15 M CaCl2 or 2.5 µg/ml tunicamycin for 0.5 and 1.5 hr, respectively. The rightmost column of the heat map shows the 91 genes containing the CDRE motif within their promoter in orange. The color bars indicate relative expression and ChIP enrichment ratios between experimental and control strains. All microarray expression and ChIP-chip experiments were performed in replicate with dye reversal. (C) The Venn diagram shows the overlap between the 339 differentially expressed genes in the transcriptome experiments and the genes identified from the ChIP-chip analysis with Prz1 promoter occupancy in the presence of CaCl2 or tunicamycin. The significance of the overlap is indicated as P-values that were determined using a hypergeometric distribution. (D) A DNA motif generated by MEME from promoter analysis of the 165 putative target genes of Prz1. This motif is similar to the CDRE motif (5′-AGCCTC-3′) previously discovered in Deng et al. (2006).