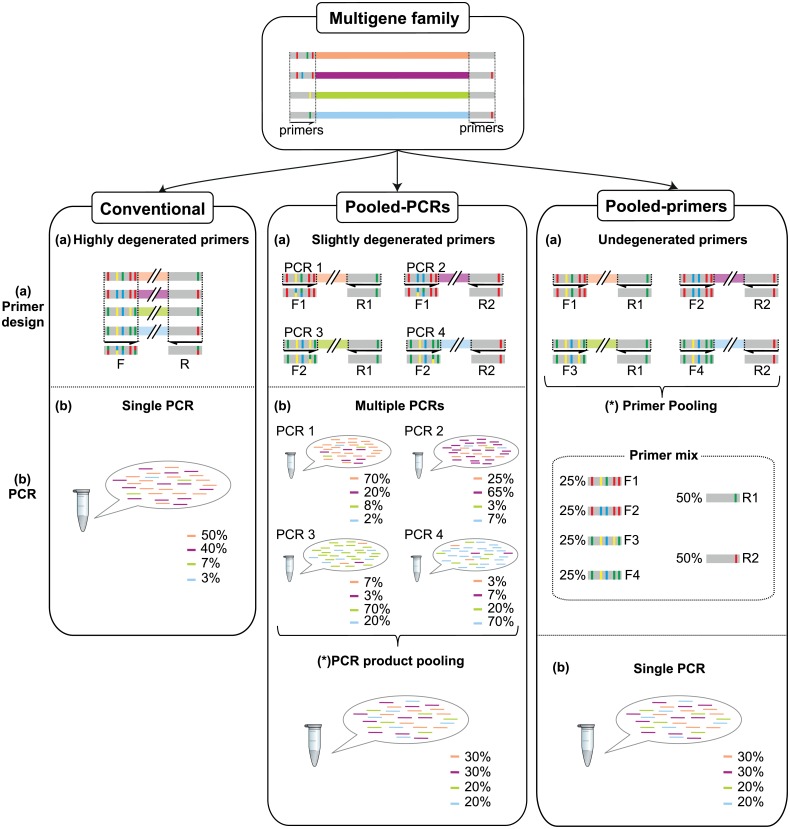
Fig 1. Diagram depicting the three alternative multigene family amplification strategies compared in this study.
When amplifying multigene families, or other complex targets, some alleles may contain variants in the priming region, here reflected by colored bars. (a) Primer design: The conventional strategy tries to match all known variation by designing highly degenerate primers, but some variants might be missed because of unknown variation or in an attempt to avoid highly-degenerate primers. Degenerate nucleotides in the primers are represented by bars with more than one color. In the pooled-PCRs strategy, allele groups with similar priming regions are targeted separately with low-degeneracy primers and by taking into account the observed phase. In the pooled-primer approach, non-degenerate primers targeting each known flanking region are pooled according to the expected number of targeted alleles. (b) PCR amplification: The conventional and pooled-primers strategies amplify all alleles in a single PCR while in the pooled-PCRs strategy an independent PCR is performed for each primer pair. Both the conventional and pooled-PCRs approaches yield unbalanced libraries, but in the case of pooled-PCRs each library is biased toward a different set of alleles. Biases in the pooled-primers approach are minimized because all alleles are primed by perfectly matching primers at the right concentration. (c) PCR yield pooling: This step is exclusive to the pooled-PCRs approach and attempts to produce a final balanced sequencing library by pooling independent PCRs taking into account the number of perfectly targeted alleles. Note that this diagram is a sketch for illustrative purposes only and does not reflect the alleles, primers or libraries used in this study.