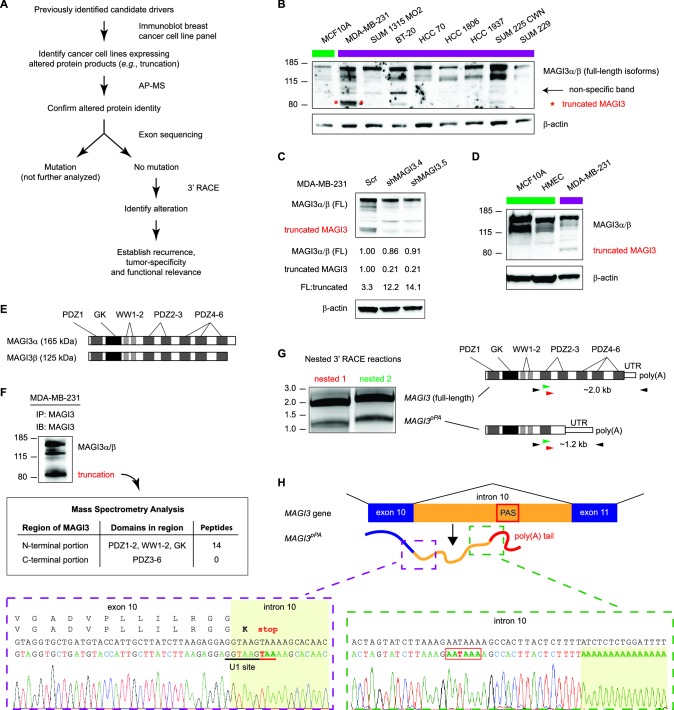
Figure 1. Premature polyadenylation of MAGI3 in the MDA-MB-231 breast cancer cell line causes the expression of a truncated MAGI3 protein.
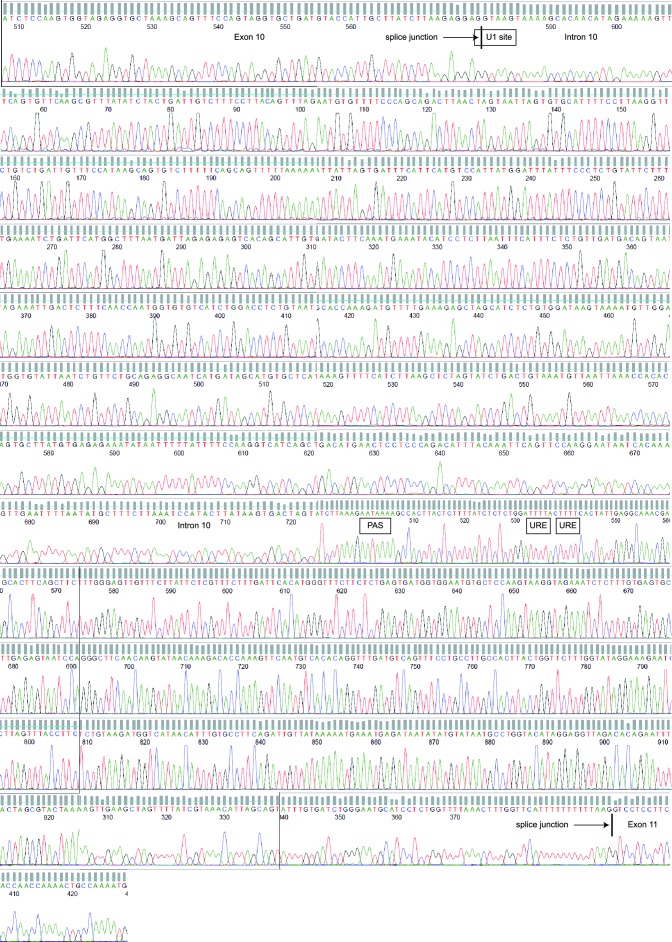
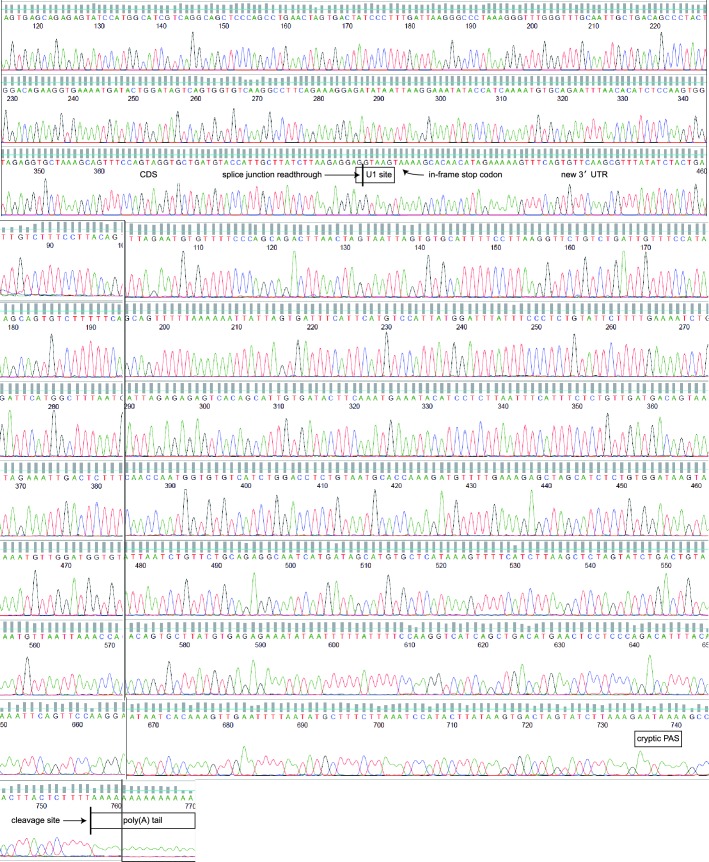
(A) Strategy for identifying protein products altered by previously overlooked mechanisms. Protein products of previously identified candidate driver genes (Ni et al., 2013) were screened by immunoblotting. Altered products were identified by electrophoretic mobility shift and confirmed by affinity purification-mass spectrometry (AP-MS). Exons of corresponding genes were sequenced. Genes not affected by coding-region DNA mutations were analyzed by 3′ RACE and sequencing to identify the nature of the altered product and the associated mechanism of alteration. Alteration events caused by mechanisms not widely appreciated in cancer were subsequently investigated for tumor-specific recurrence and functional relevance. (B) Immunoblot of MAGI3 gene products in the screening panel of human breast cell lines. Non-transformed (green) and transformed (violet) cell lines are indicated. A truncated MAGI3 protein (indicated by red asterisks) is expressed in MDA-MB-231 cells. Immunoblot of β-actin is included to show loading. Approximate molecular mass markers are indicated in kDa. (C) Immunoblots of MAGI3 gene products and β-actin levels in MDA-MB-231 cells transduced with the indicated shRNA constructs. The relative levels of full-length and truncated MAGI3 proteins following RNAi-mediated depletion are normalized to β-actin levels. The ratios of full-length to truncated MAGI3 (FL:truncated) are also quantified. (D) Immunoblot of MAGI3 gene products in the indicated non-transformed cell lines (green) and MDA-MB-231 breast cancer cell line (violet). Immunoblot of β-actin is included to show loading. Approximate molecular mass markers are indicated in kDa. (E) Diagrams of the full-length MAGI3 protein isoforms α and β. Both full-length isoforms possess the indicated arrangement of 6 PDZ domains (dark grey), 2 WW domains (light grey) and a catalytically inactive GK domain with homology to the yeast guanylate kinase (black). In addition, isoform α has an extended C-terminal sequence with no known function. (F) Immunoblot for three MAGI3 protein isoforms immunoprecipitated from MDA-MB-231 cell lysate. The gel slice containing the smallest, truncated MAGI3 protein was analyzed by mass spectrometry, and peptides were mapped exclusively to the N-terminal half of the protein. Approximate molecular mass markers are indicated in kDa. (G) Detection of full-length and truncated MAGI3 mRNA isoforms by 3′ RACE. Products from two independent nested 3′ RACE reactions performed on MDA-MB-231 RNA were separated by agarose gel electrophoresis. Truncated and full-length MAGI3 transcripts are indicated. Diagrams show regions targeted for amplification and locations of primary (black) and nested (green/red) primers used for 3′ RACE. (H) Diagram of the truncated and prematurely polyadenylated MAGI3 mRNA (MAGI3pPA) generated from the MAGI3 gene locus despite the absence of genetic mutations within the gene. Green box: 3′ RACE sequencing results of MAGI3pPA show premature cleavage and polyadenylation (shaded yellow) following a cryptic PAS (outlined in red) in intron 10. The corresponding intron 10 genomic sequence is shown above in black. Violet box: Upstream of the pPA event, 3′ RACE sequencing results show a short extension of the open-reading frame into intron 10. The corresponding exon 10 and intron 10 genomic sequence is shown above in black, with the intact U1 snRNA binding site (GGTAAG) underlined in black. Intron 10 is shaded yellow. A stop codon (red underscore) occurs 6 nt following the exon-intron junction. Encoded amino acid sequence is shown in black above the nucleotide sequence (wild-type MAGI3, upper line; MAGI3pPA, lower line).